-
Notifications
You must be signed in to change notification settings - Fork 16
Measure_Nuclei_And_Membranes_Tool
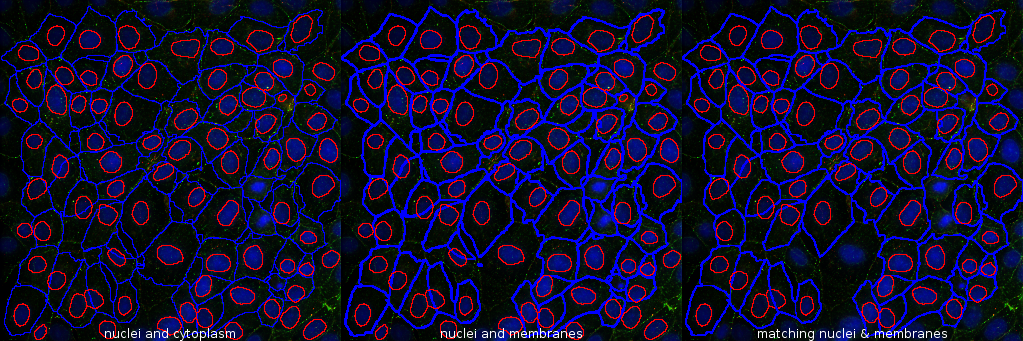
The tool takes the results of cell and nuclei segmentation done with cellpose [1] and transforms the cytoplasm selection into a membrane selections, removes rois for which one membrane does not match one nucleus and measures the membranes and nuclei in all channels but the nuclei-channel.
To install the tool save the file measure_nuclei_and_membranes.ijm into the folder macros/toolsets of your FIJI installation.
Select the "measure_nuclei_and_membranes" toolset from the >> button of the ImageJ launcher.
- the first button opens this help page
- the
o-button opens a batch script that converts cellpose results_cp_outlines.txtto rois in the overlays of the input images - the
c-button transforms the cytoplasm selections into membrane selections - the
m-button removes rois for which one membrane does not match one nucleus and measures the membranes and nuclei
Use our modified cellpose version to batch segment in two runs the nuclei and the cytoplasms of the cells and save the results into the same output folder. Press the o-button of the toolset and run the python-script from the editor to convert the cellpose outlines to ImageJ rois. The rois will be stored in the overlays of the input images.
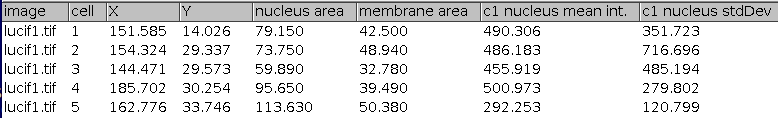
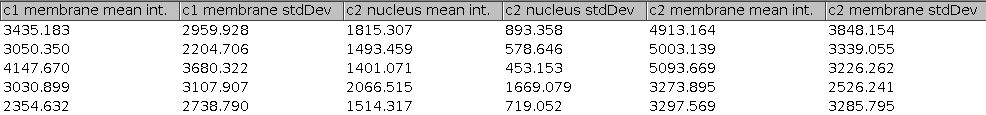
Press the c-button to transform the cytoplasm rois into membrane rois. Finally press the m-button to remove non-matching nucleus/membrane rois and measure the intensities in the membranes and nuclei in all channels but the nuclei channel.
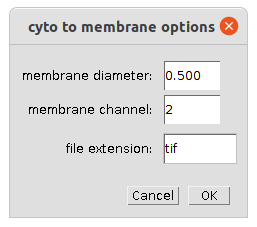
Right-click the c-button to open the options for the cytoplasm to membrane transformation
- membrane diameter - the diameter of the membrane band-like rois
- membrane channel - the channel containing the cell (cytoplasm) staining
- file extension - the file extension of the input image files without the point, for example
tif
- nuclei channel - the channel containing nuclei, the intensities in this channel will not be measured
- file extension - the file extension of the input image files without the point, for example
tif
[1] Stringer, C., Wang, T., Michaelos, M. et al. Cellpose: a generalist algorithm for cellular segmentation. Nat Methods 18, 100–106 (2021). https://doi.org/10.1038/s41592-020-01018-x

 Volker Bäcker
Volker Bäcker