-
Notifications
You must be signed in to change notification settings - Fork 16
Incucyte_Exporter
Export the images from the Incucyte. Stitch the images, create a time-series and align the frames.
The source code in git-hub can be found here.
You need to have the HyperStackReg plugin installed, see https://github.com/ved-sharma/HyperStackReg for details on the plugin. Please install the version from the first link above, by copying the jar-file into the folder plugins of your FIJI installation. This version is compiled for java 1.8. The HyperStackReg-plugin also needs the TurboReg-plugin, which can be installed via the BIG-EPFL update site.
To install the tool save the file incucyte_exporter.ijm into the folder macros/toolsets of your FIJI installation.
Select the "incucyte_exporter" toolset from the >> button of the ImageJ launcher.
- ?
- Open this help page.
- export raw images as std. tif
- Convert the images to a standard tif-format.
- stitch images
- Calculate the stitchings for the images.
- clean images
- Normalize the intensities and apply a rolling ball background subtraction.
- merge images
- Create the mosaic.
- mark empty images
- Prefixes the file name of the well-image with ``Empty_`` if no object has been found in the well.
- make time series
- Create a time series in one file from the different images of a well.
- batch export images
- Run all the before mentioned steps on the images.
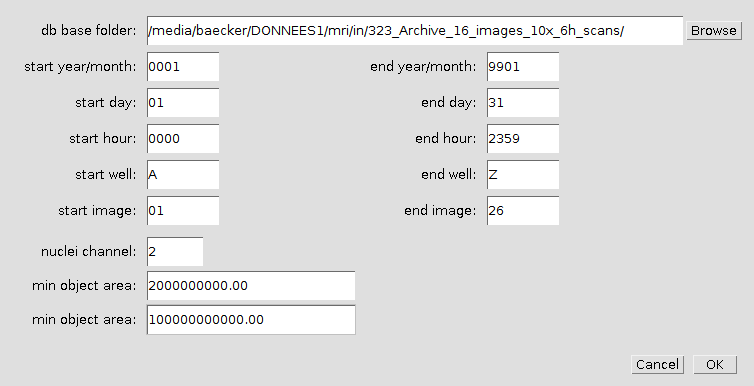
The first time you run one of the steps, you will be asked for the base-folder of the image database. This is the folder that contains the subfolder EssenFiles. A right click on one of the buttons opens the options dialog. The options dialog allows to change the image database, to limit the wells that will be exported and to specify the conditions under which a well is considered empty.
You can use the buttons from left to right to run the consecutive steps, or you can use the b-button to run all steps automatically. At the end you will find a mosaic-time-series-file per well.
Right-click on any of the buttons to open the options-dialog.
- db base folder
- The base folder of the image database (contains a folder 'EssenFiles').
- start year/month, end year/month
- Restrict the images to be exported to those between start year/month and end year/month. The first two digits are for the year and the second two for the month: yymm. The default values mean January 2000 to January 2099.
- start day, end day
- Restrict the images to be exported to those between start day and end day.
- start hour, end hour
- Restrict the images to be exported to those between start hour and end hour.
- start well, end well
- Restrict the images to be exported to those in the wells between start well and end well. This options is only used by the first step. The following steps will use all images exported by the first step.
- start image, end image
- Restrict the images to be exported to those between start image and end image. This options is only used by the first step. The following steps will use all images exported by the first step.
- nuclei channel
- The number of the channel that is used to calculate the stitching and to decide whether a well is empty.
- min. object area
- The minimal area that must be detected so that the well is not considered empty.
- max. object area
- Bigger objects are not taken into account in the decision whether a well is empty or not.
- The output images will be written into the
ScanData-folder - Running HyperStackReg again on the result can help to get better aligned timepoints

 Volker Bäcker
Volker Bäcker