-
Notifications
You must be signed in to change notification settings - Fork 13
Beat Detection
Beat detection is accomplished using a correlation-based template matching algorithm. A sliding window is passed across the ECG signal, and a correlation coefficient, rxy, is computed between the template and the ECG contained within each window of equal length. Here the correlation coefficient represents the similarity between template and ECG segment. This process produces a new data series containing correlation coefficients rxy. The correlation coefficient between two N length segments of x and y is computed using the following equation:
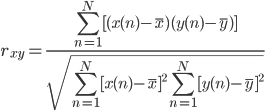
If the correlation, rxy, between template and ECG segment exceed a defined upper threshold value a detection flag is set to high (Figure.1-c). This flag is held in a high state until rxy drops below a defined lower threshold (Figure.1-d). A beat is set as the largest rxy peak between any high and low detection flag events (Figure.1-b). Detected beats are marked with green circles. This double threshold method helps eliminate multiple peak detections within a close vicinity of one another.
Computational speed of template matching can be improved using the following three methods (see the function detectBeats() within ecgViewer.m and see matchTemplate.m ):
-
Skip Indexing - This method involves reducing the number of times the sliding window translates across the ECG signal. By translating every third ECG sample instead of indexing one sample, 1/3 as many correlation coefficients are computed. Skipped rxy values are replaced using cubic spline interpolation.
-
Parallelization - This method involves parallelizing a portion of the template matching code for multi-core processor environments.
-
Combination - This method combines both the skip indexing and parallelized methods. Skipping samples, parallelizing code, and a combination of the two methods reduce computation time (17.2 s original) by approximately 64% (6.2 s), 58% (7.3 s), and 82% (3.16 s) respectively. These data are based on the results of 3 simulations of each technique using an ECG segment of 600,000 samples and a template of 161 samples in length.
Two types of detection methods exist on the ECG Viewer GUI:
-
Template Matching – standard template matching
-
Self Template – This methods using the defined template file to create a new template base on the current ECG file. The current template is used to detect QRS complexes from the first 60,000 samples of ECG. These detected beats are then averaged together to create a “self template”. This self template is used in beat detection. NOTE: The sample length and template width may need to be modified for your preference.
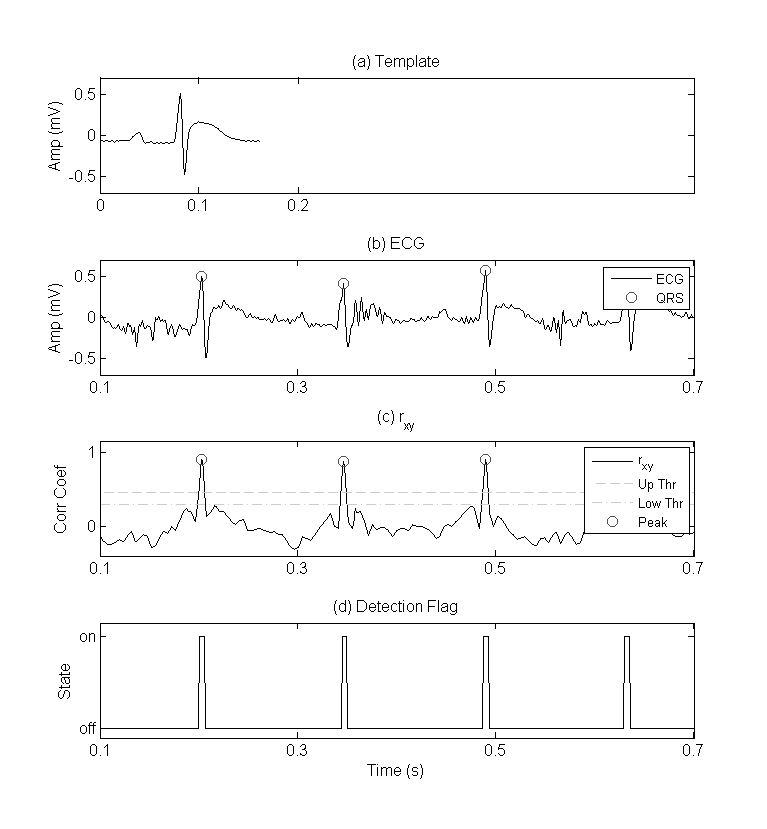
Figure 1 – QRS detection using template matching. Plots include (a) example template used for QRS detection by template matching, (b) rat ECG with detected QRS complexes, (c) rxy data series and peaks detected using the double threshold technique, and (d) state of the detection flag used in the double threshold technique.
- ECG Viewer’s template matching marks beats as the center of a matched template. For example, if you make the center of your template correspond with the R-wave peak, then template matching will mark R-waves on your ECG data, etc.
- Averaging several QRS templates together may help create a more universal template
- elaborate on building a template