-
Notifications
You must be signed in to change notification settings - Fork 4
JSON EffectBlocks
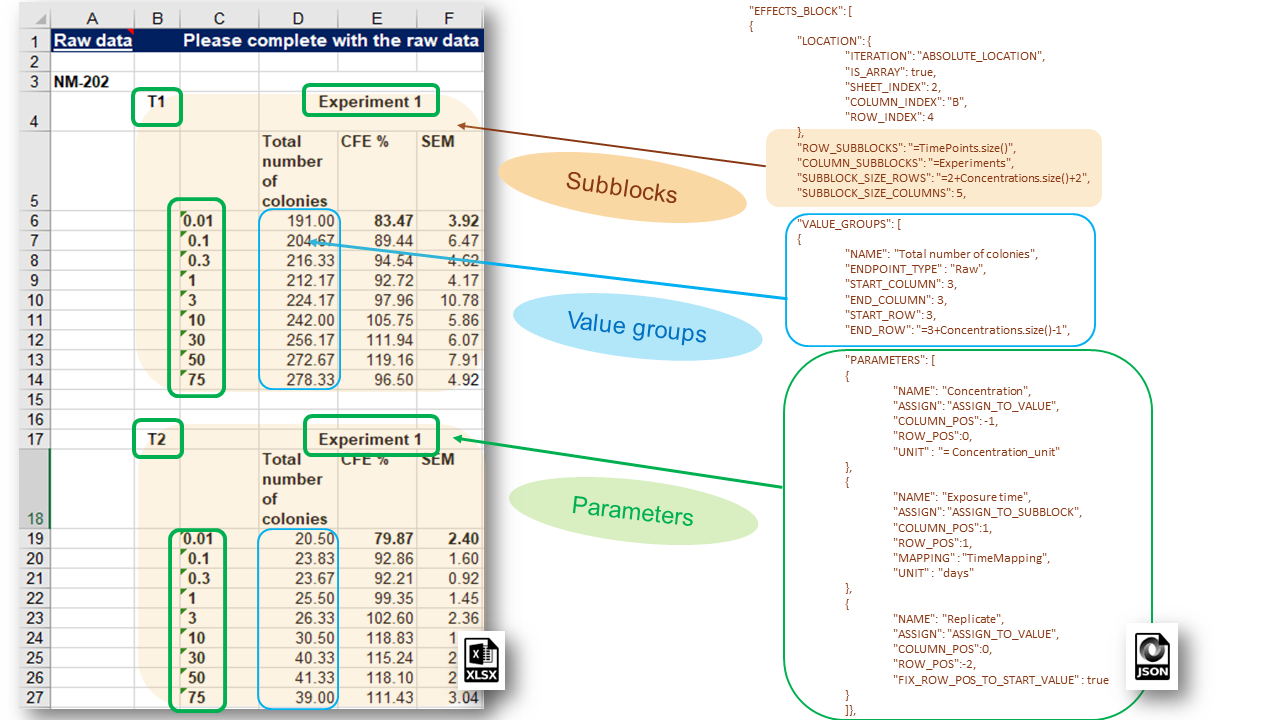
More complex data organizations of the experimental measurements stored in spreadsheet files are also often used from the nanosafety community. The typical bioassay experiment includes measurements with variation of several experimental factors (the latter yields combinations of experimental conditions and multiple measurements), as in the dose response data recorded in the IOM Excel templates illustrated in the figure below:
The CFE (Colony Forming Efficiency) assay is performed using 2 replicates, 2 different timings and 9 concentrations of silicon dioxide (i.e. 2 x 2 x 9 = 36 measurements must be reported is full experiment is performed). With some effort, it would be possible to handle this layout using the EFFECT section described above, but it would be very inefficient and prone to human errors. The NMDataParser functionality was extended to support blocks of effects to avoid enumerating each combination of the experimental conditions values by hand. A new JSON section EFFECT_BLOCKS is introduced within PROTOCOL_APPLICATIONS objects. It is used to configure a simultaneous reading of many effects, grouped in blocks of measurements according to the variations of the experimental factors. In this way one EFFECT_BLOCK can describe multiple values found on one sheet. This feature is crucial for configuration of HTS (High-throughput screening) data import which is practically impossible to handle each single value by hand.
NMDataParser supports several levels of block data aggregation which allows very complex spreadsheet data organizations to be mapped onto the eNanoMapper data model. Each block can be divided into rectangular grid of sub-blocks. Each sub-block can contain several value groups. Each value group is a set of measurements where each measurement is associated with a list of experimental conditions called in this context parameters of the value group (not to be confused with the parameters of the Protocol Applications).
EFFECTS_BLOCK attribute is a single JSON object or an array. This means that it may be defined with JSON syntax:
"EFFECTS_BLOCK": [
{
...
}.
{
...
}
]
as well as with syntax:
"EFFECTS_BLOCK":{
...
}
Each EFFECTS BLOCK is defined by its LOCATION attribute configured as an EDL object and several attributes to define the sub-blocks grid:
| JSON attribute | meaning |
|---|---|
| ROW_SUBBLOCKS | number of individual sub-blocks in the main effect block in a row (line) |
| COLUMN_SUBBLOCKS | number of individual sub-blocks in the main effect block in a column |
| SUBBLOCK_SIZE_ROWS | number of rows contained in one sub-block |
| SUBBLOCK_SIZE_COLUMNS | number of columns contained in one sub-block |
EFFECTS BLOCK also includes an array section VALUE_GROUPS. For a correct configuration of the sub-blocks, the same number of rows and columns in each sub-block and distances (column/row shifts and padding) between them are needed i.e. correct spreadsheet generation is mandatory. Section Effect Blocks Advanced describes in detail the advanced utilization of effect blocks. For the purpose of efficient configuration of complex data organizations in effect blocks, the JSON syntax for handling of variables is introduced.
- Home
- Quick start
- Data templates
- eNanoMapper Data Model
- Parser configuration via JSON
- Available templates
- How to
- Additional information