-
Notifications
You must be signed in to change notification settings - Fork 199
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Gff3tabix representation not informative or absent #1103
Comments
|
@thomasvangurp do you have any additional information about this? I am not sure I am able to reproduce this issue. Did you make a new track using GFF3Tabix in the second picture that was based on the NCList data in the first picture? Also what JBrowse version are you using? Note that JBrowse 1.14.1 added better histograms for GFF3Tabix (see release-notes and #1039) |
|
@thomasvangurp any update on this? is there a sample data file that helps recreate this issue? |
|
@cmdcolin Sorry for the delay, i made the new config using gff3tabix, this is entry (without the "style" and "menutemplate" option: {
"key" : "PGSC DM v4.03 gene models",
"trackType" : "CanvasFeatures",
"storeClass" : "JBrowse/Store/SeqFeature/GFF3Tabix",
"maxFeatureScreenDensity" : 0.05,
"urlTemplate" : "solgenomics_tracks/gene_models/PGSC_DM_V403_genes.gff3.gz",
"tbiurlTemplate" : "solgenomics_tracks/gene_models/PGSC_DM_V403_genes.gff3.gz.tbi",
"storeTimeout" : 10000,
"metadata" : {
"category" : "solgenomics/Gene models",
"description" : "Gene models from PGSC for S. tuberosum Group Phureja DM1-3 Genome Annotation v3.4 mapped to v4.03"
},
"type" : "CanvasFeatures",
"label" : "gene_models"
},I am running Jbrowse 1.14.2. You can download the potato genome here: http://solanaceae.plantbiology.msu.edu/data/potato_dm_v404_all_pm_un.fasta.zip The tabix indexed gff3.gz are attached (note that the .tbi file has the .txt extension attached as uploading a .tbi file is not permitted by github): PGSC_DM_V403_genes.gff3.gz.tbi.txt please let me know if you can reproduce the issue with the data attached. EDIT: {
"maxFeatureSizeForUnderlyingRefSeq": 250000,
"subfeatureDetailLevel": 2,
"maxFeatureScreenDensity": 0.05,
"enableCollapsedMouseover": false,
"disableCollapsedClick": false,
"maxFeatureGlyphExpansion": 500,
"maxHeight": 600,
"histograms": {
"description": "feature density",
"min": 0,
"height": 100,
"color": "goldenrod",
"clip_marker_color": "red"
},
"style": {
"_defaultHistScale": 4,
"_defaultLabelScale": 30,
"_defaultDescriptionScale": 120,
"showLabels": true,
"showTooltips": true,
"label": "id",
"description": "name",
"className": "feature",
"linkTemplate": "http://solgenomics.net/search/quick?term={id}"
},
"displayMode": "normal",
"events": {},
"menuTemplate": [
{
"label": "View details",
"title": "{type} {name}",
"action": "contentDialog",
"iconClass": "dijitIconTask"
},
{
"label": "Highlight this gene",
"iconClass": "dijitIconConnector"
},
{
"label": "Ask SGN about {id}",
"action": "newWindow",
"iconClass": "dijitIconDatabase",
"url": "http://solgenomics.net/search/quick?term={id}"
},
{
"iconClass": "dijitIconDatabase",
"action": "newWindow",
"url": "http://www.ebi.ac.uk/ebisearch/search.ebi?db=proteinSequences&t={id}",
"label": "Ask EMBL about {id}"
},
{
"iconClass": "dijitIconDatabase",
"action": "newWindow",
"url": "http://www.google.com/search?q={id}",
"label": "Ask Google about {id}"
}
],
"key": "PGSC DM v4.03 gene models",
"trackType": "CanvasFeatures",
"storeClass": "JBrowse/Store/SeqFeature/GFF3Tabix",
"urlTemplate": "solgenomics_tracks/gene_models/PGSC_DM_V403_genes.gff3.gz",
"tbiurlTemplate": "solgenomics_tracks/gene_models/PGSC_DM_V403_genes.gff3.gz.tbi",
"storeTimeout": 10000,
"metadata": {
"category": "solgenomics/Gene models",
"description": "Gene models from PGSC for S. tuberosum Group Phureja DM1-3 Genome Annotation v3.4 mapped to v4.03"
},
"type": "JBrowse/View/Track/CanvasFeatures",
"label": "gene_models",
"baseUrl": "http://my.url/data/Solanum_tuberosum_v4.04/existing_tracks/",
"index": 2
} |
|
Should be fixed by #1123 ! Thanks so much for the good sample data and patience!!! |
I have loaded several tracks using a tabix-indexed bgzipped GFF3 file in a genome browser (not publicly accesible :-( ). The feature density display is not workable for me currently. When I compare a feature density display for the same track data comparing
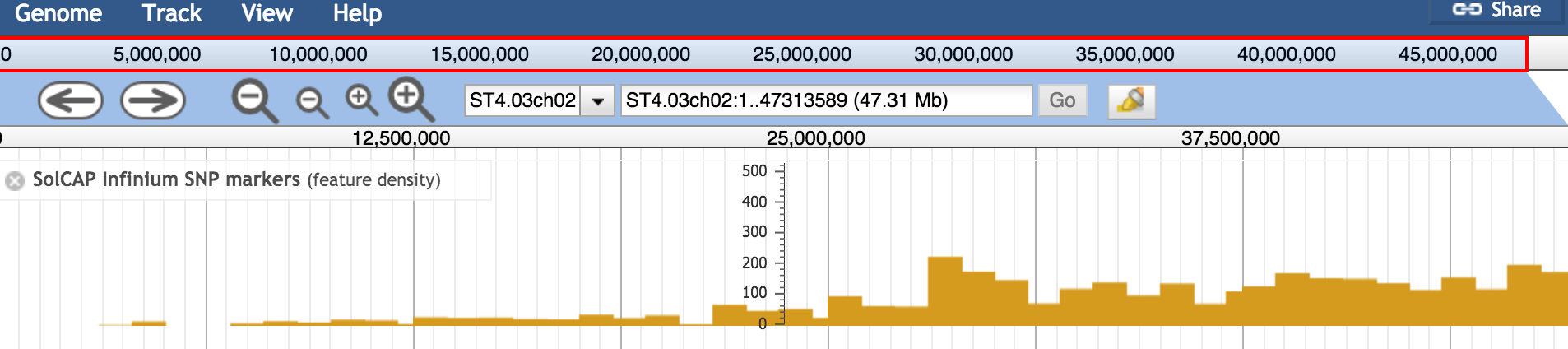
"storeClass": "JBrowse/Store/SeqFeature/NCList",and"storeClass": "JBrowse/Store/SeqFeature/GFF3Tabix",the display for the NClist is working: see solgenomics jbrowsewheras the gff3tabix track displays the following:

Zoomed in both tracks work as they should. For the gene tracks I do get a display of some features, but is is very sparse:
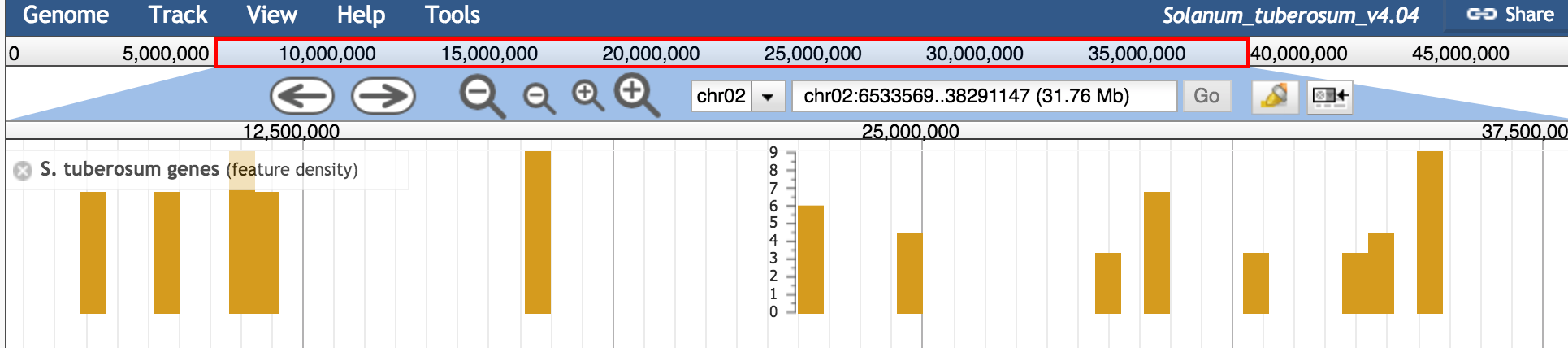
Could is be that the interval size of the tbi files is simply to big to be usefull for displaying a rich and detailed feature density plot? If that's the case, would csi indexes solve this issue? see my comment in #1086
The text was updated successfully, but these errors were encountered: