If you want to watch a 2-Minute video introduction, click the image below:
rixpress streamlines creation of micropipelines (small-to-medium,
single–machine analytic pipelines) by expressing a pipeline in idiomatic
R while delegating build orchestration, dependency management, and
multi-language execution to the ‘Nix’ build system. It is inspired by
the user experience of the {targets} package and builds on the {rix}
package to obtain fully reproducible development/runtime environments.
Key ideas:
- Define pipeline derivations with concise
rxp_*()helper functions. - Seamlessly mix R, Python, Julia, Quarto / Markdown steps.
- Reuse hermetic environments (pinned system + language dependencies)
defined via
{rix}and adefault.nix. - Visualize and inspect the DAG; selectively read, load, or copy outputs.
- Export/import build artifacts to speed up CI or share results.
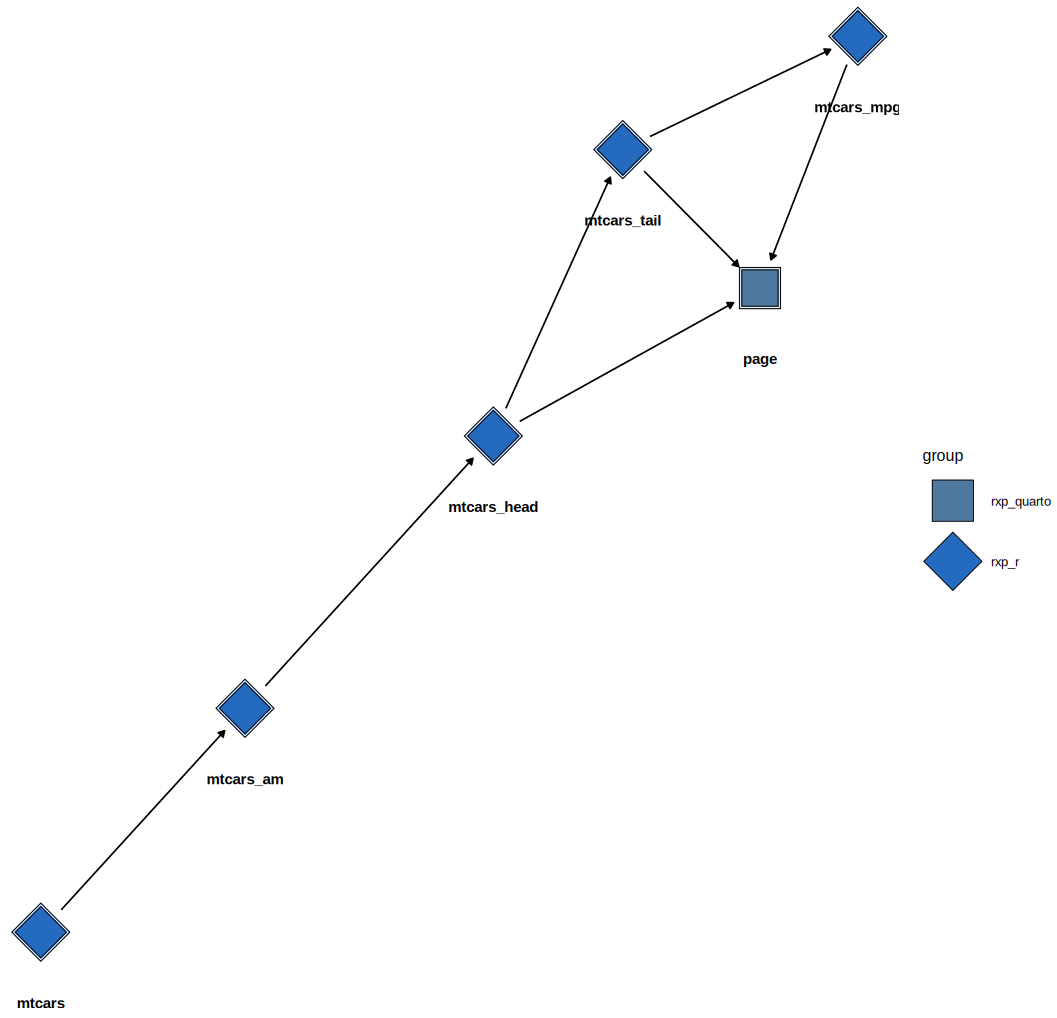
Here is what a basic pipeline looks like:
library(rixpress)
list(
rxp_r_file(
mtcars,
'mtcars.csv',
\(x) (read.csv(file = x, sep = "|"))
),
rxp_r(
mtcars_am,
filter(mtcars, am == 1)
),
rxp_r(
mtcars_head,
head(mtcars_am)
),
rxp_r(
mtcars_tail,
tail(mtcars_head)
),
rxp_r(
mtcars_mpg,
select(mtcars_tail, mpg)
),
rxp_qmd(
page,
"page.qmd"
)
) |>
rxp_populate()- Motivation
- Feature Highlights
- Examples
- Getting started
- Visualization
- Exporting / Importing Artifacts
- Contributing
- Scope
- Acknowledgements
Reproducibility involves two intertwined concerns: (1) the environment (system libraries, compilers, language interpreters, packages) and (2) the execution graph of analytic steps with their inputs and outputs. There are many tools to address both of these concerns, but orchestrating them is not always easy.
Another developing trend, in my opinion, is that data science is becoming increasingly polyglot: teams rarely restrict themselves to a single language. Python dominates machine learning, R excels at statistical modeling and visualization, and Julia offers high-performance numerics with a syntax that feels familiar to both communities. Analysts and researchers often need to combine strengths from all three, moving data and results fluidly across tools.
This trend makes a unifying foundation essential. Without it, people waste time stitching together environments, dealing with dependency conflicts, or struggling to reproduce results on different machines. Nix provides that foundation by declaratively describing environments which include system libraries, compilers, interpreters, and packages in a way that is reproducible down to exact versions and build instructions.
The {rix} package brings this power into the R ecosystem. It makes it
easy to generate declarative, date-pinned Nix expressions that work
consistently across systems. This expressions can then be used to build
reproducible development environments that include programming
languages, packages and other tools. {rixpress} addresses the next
part, the execution graph. With a concise R API, it lets users describe
analytic steps, inputs, and outputs, while delegating execution to Nix
for guaranteed determinism.
Together, this means data scientists can orchestrate polyglot pipelines that combine R, Python, and Julia seamlessly, with environments and workflows that are portable, reproducible, and future-proof.
If you are interested in a Python-first port of {rixpress}, check out
ryxpress.
- Uniform
rxp_*()constructors for files, R functions, Python / Julia code, and Quarto documents. - Cross-language object exchange via serialization helpers.
- Pipelines as compositions of pure functions: derivations are content-addressed and cached in the Nix store.
- DAG visualization (e.g.
rxp_ggdag(),rxp_visnetwork()). - Target introspection (
rxp_read(),rxp_load(),rxp_copy(),rxp_trace()). - Artifact export/import for CI acceleration.
Here is what a basic pipeline looks like:
library(rixpress)
list(
rxp_r_file(
mtcars,
'mtcars.csv',
\(x) (read.csv(file = x, sep = "|"))
),
rxp_r(
mtcars_am,
filter(mtcars, am == 1)
),
rxp_r(
mtcars_head,
head(mtcars_am)
),
rxp_r(
mtcars_tail,
tail(mtcars_head)
),
rxp_r(
mtcars_mpg,
select(mtcars_tail, mpg)
),
rxp_qmd(
page,
"page.qmd"
)
) |>
rxp_populate()Running rxp_populate() generates a pipeline.nix file, which contains
the build instructions for all derivations and final outputs expressed
as Nix code. You can define derivations that run Python or Julia code,
and objects can be exchanged between R and Python by using rxp_py2r()
and rxp_r2py(), or by serializing to a common format such as JSON. By
default, calling rxp_populate() also builds the pipeline, but it’s
possible to only generate the pipeline.nix file and build the pipeline
later using:
rxp_make()The build process assumes the presence of a default.nix file that
defines the computational environment the pipeline runs in; this file
can be generated with the {rix} package. The default.nix typically
defines an environment with R and required R packages (and optionally
Python/Julia and their packages), Quarto, and any necessary system-level
dependencies pinned to a specific date to ensure reproducibility.
In the example above, the first derivation reads mtcars.csv (in the
example it’s pipe-separated, i.e. a .psv file). Each output (for
example, mtcars, mtcars_am, mtcars_head, mtcars_tail,
mtcars_mpg, page) is built by Nix within the environment defined by
default.nix. Concretely, {rix} makes using Nix as a package manager
easier for R users, and {rixpress} makes it easy to use Nix as a build
automation tool.
And this is what a polyglot pipeline, using both R and Python, looks like:
library(rixpress)
list(
rxp_py_file(
name = mtcars_pl,
path = "data/mtcars.csv",
read_function = "lambda x: polars.read_csv(x, separator='|')"
),
rxp_py(
name = mtcars_pl_am,
expr = "mtcars_pl.filter(polars.col('am') == 1)",
user_functions = "functions.py",
encoder = "serialize_to_json",
),
rxp_r(
name = mtcars_head,
expr = my_head(mtcars_pl_am),
user_functions = "functions.R",
decoder = "jsonlite::fromJSON"
),
rxp_r(
name = mtcars_mpg,
expr = dplyr::select(mtcars_head, mpg)
)
) |>
rxp_populate(project_path = ".", build = FALSE)Because the pipeline is built using Nix, outputs are stored in the Nix
store under /nix/store/. To make working with these outputs easier,
{rixpress} provides several helper functions:
rxp_read("mtcars_mpg")— read the content ofmtcars_mpginto R (the return value depends on the derivation type: an R object, a file path, etc.);rxp_load("mtcars_mpg")— load objects from the result into the global environment;rxp_copy("page")— copy outputs (e.g. a generated document) from the Nix store into the current working directory so you can open or inspect them there.
Python objects will be converted into their equivalent R objects if
{reticulate} is available in the environment. For complex outputs such
as documents (for example the Quarto document page above),
rxp_read("page") returns the output file path; you can then open it
with browseURL() or copy it into your working directory with
rxp_copy().
{rixpress} is flexible; please consult the examples repository for
many different patterns and complete demos:
https://github.com/b-rodrigues/rixpress_demos/tree/master
While {rixpress} is a regular R package, there is little point in
using it without having Nix installed. If you are not familiar with Nix,
I recommend you first start by checking out {rix}, which will teach
you how to use Nix to set up reproducible development environments for
your R projects!
We recommend you check out the following vignettes to get started:
It is possible to visually inspect the pipeline:
# ggdag-based static plot
rxp_ggdag()DAG
# Interactive network (visNetwork)
rxp_visnetwork()To create an artifact suitable for CI, use:
rxp_dag_for_ci()This will export the directed acyclic graph of the pipeline as a dot
file, making it possible to visualize in CI (using
stacked-dag for
example).
Speed up continuous integration or move a cache between machines:
# Export build products
rxp_export_artifacts()
# Import elsewhere before building
rxp_import_artifacts()Pull requests are welcome. If you’re unsure whether to open one, feel free to open an issue first to discuss your idea. For contributor guidelines, see CONTRIBUTING.md.
If you plan to contribute documentation or vignettes, please:
- Add runnable, minimal examples.
- Prefer small datasets and short-running examples.
- Document required system dependencies for the example (through a
default.nix).
Please note that this package is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
Please refer to the vignette("scope") to learn more about what
{rixpress} will and will not support.
- Inspired by the design and ergonomics of
{targets}. - Built atop the reproducibility foundations provided by
{rix}and the ‘Nix’ ecosystem. - rOpenSci peer review contributors and reviewers for feedback that shaped the interface.