scikit-na is a Python package for missing data (NA) analysis. The package includes many functions for statistical analysis, modeling, and data visualization. The latter is done using two packages — matplotlib and Altair.
- Interactive report (based on ipywidgets)
- Descriptive statistics
- Regression modeling
- Hypotheses tests
- Data visualization
If you find this package useful, please consider donating any amount of money. This will help me spend more time on supporting open-source software.
Package can be installed from PyPi:
pip install scikit-naor from this repo:
pip install git+https://github.com/maximtrp/scikit-na.gitWe will use Titanic dataset (from Kaggle) that contains NA values in three columns: Age, Cabin, and Embarked.
By default, summary() function returns the results for each column.
import scikit_na as na
import pandas as pd
data = pd.read_csv('titanic_dataset.csv')
# Excluding three columns without NA to fit the table here
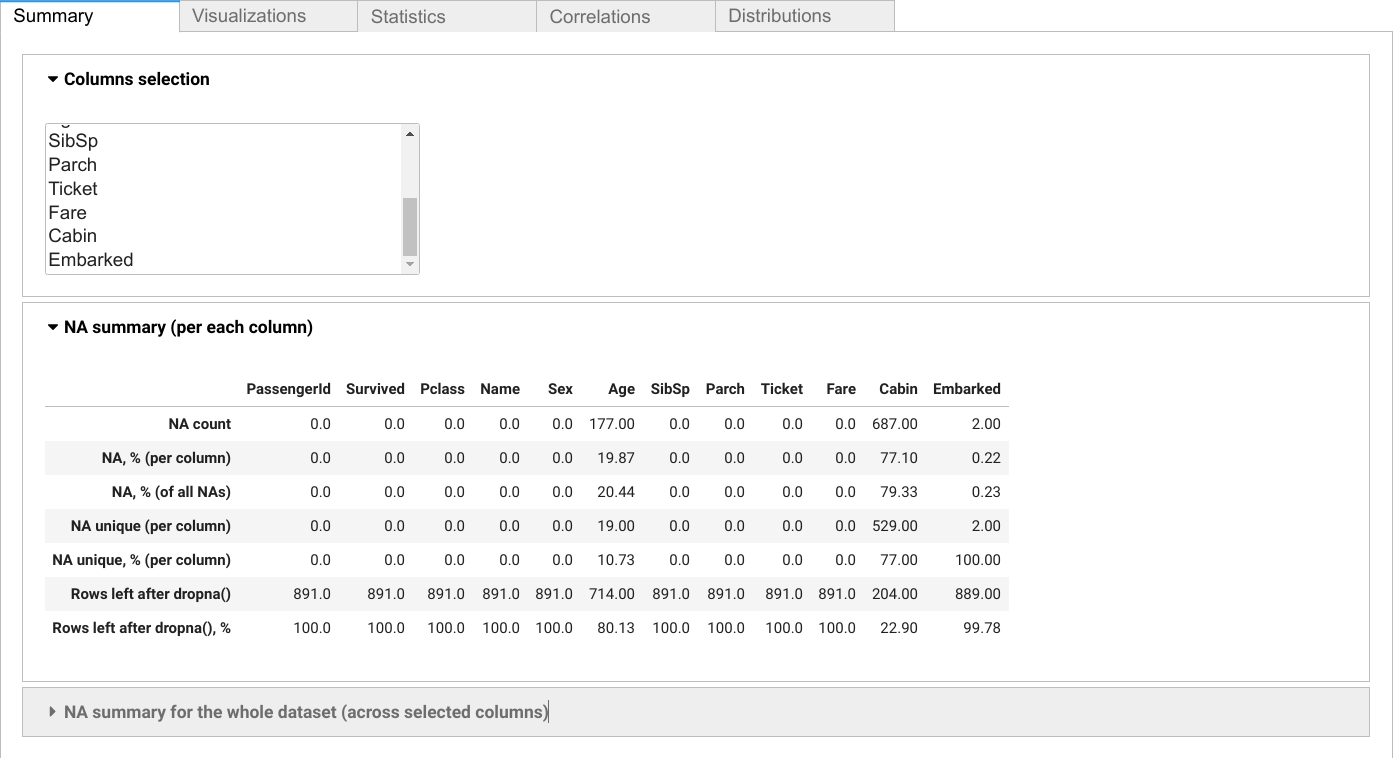
na.summary(data, columns=data.columns.difference(['SibSp', 'Parch', 'Ticket']))| Age | Cabin | Embarked | Fare | Name | PassengerId | Pclass | Sex | Survived | |
|---|---|---|---|---|---|---|---|---|---|
| NA count | 177 | 687 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| NA, % (per column) | 19.87 | 77.1 | 0.22 | 0 | 0 | 0 | 0 | 0 | 0 |
| NA, % (of all NAs) | 20.44 | 79.33 | 0.23 | 0 | 0 | 0 | 0 | 0 | 0 |
| NA unique (per column) | 19 | 529 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| NA unique, % (per column) | 10.73 | 77 | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Rows left after dropna() | 714 | 204 | 889 | 891 | 891 | 891 | 891 | 891 | 891 |
| Rows left after dropna(), % | 80.13 | 22.9 | 99.78 | 100 | 100 | 100 | 100 | 100 | 100 |
NA unique is the number of NA values per each column that are unique for it, i.e. do not intersect with NA values in the other columns (or that will remain in dataset if we drop NA values in the other columns).
We can also get a summary of missing data for the whole dataset:
na.summary(data, per_column=False)| dataset | |
|---|---|
| Total columns | 12 |
| Total rows | 891 |
| Rows with NA | 708 |
| Rows without NA | 183 |
| Total cells | 10692 |
| Cells with NA | 866 |
| Cells with NA, % | 8.1 |
| Cells with non-missing data | 9826 |
| Cells with non-missing data, % | 91.9 |
To calculate correlations between columns in terms of missing data, just call
correlate() function with your DataFrame as the first argument:
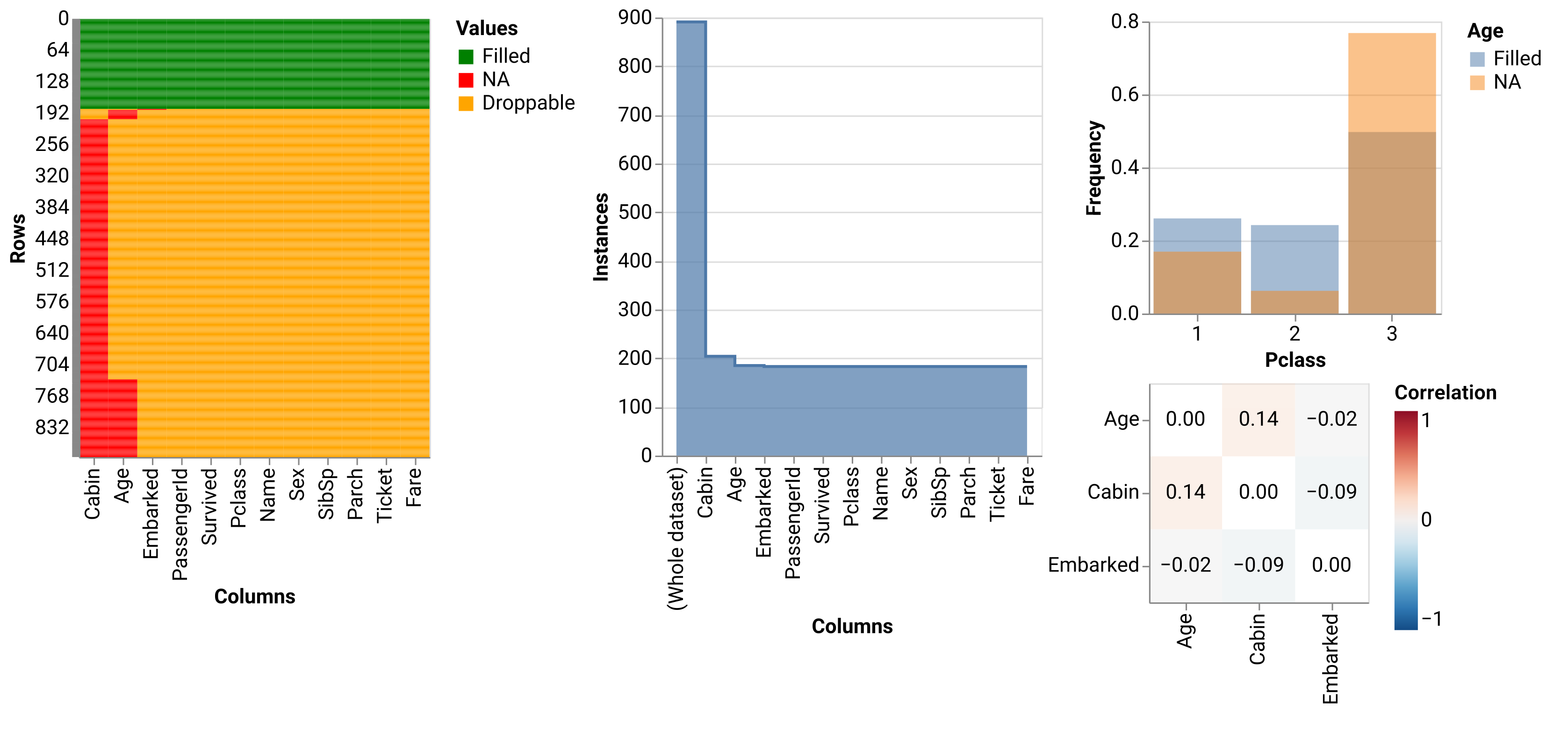
na.correlate(data, method="spearman").round(3)| Embarked | Age | Cabin | |
|---|---|---|---|
| Embarked | 1 | -0.024 | -0.087 |
| Age | -0.024 | 1 | 0.144 |
| Cabin | -0.087 | 0.144 | 1 |
This method can be used to uncover hidden patterns in missing data across many columns in a dataset. Columns with no missing data are automatically excluded.
There is a function to visualize correlations with a heatmap:
na.altair\
.plot_corr(data, corr_kws={'method': 'spearman'})
.properties(width=150, height=150)Now, let's visualize NA values on a heatmap. We will be using Altair + Vega backend:
na.altair.plot_heatmap(data)Droppables are those values that will be dropped if we simply use
pandas.DataFrame.dropna() on the whole dataset.
Stairs plot is one more useful visualization of dataset shrinkage on applying
pandas.Series.dropna() method to each column sequentially (sorted by the
number of NA values, by default):
na.altair.plot_stairs(data)After dropping all NAs in Cabin column, we are left with 21 more NAs (in Age
and Embarked columns). This plot also shows tooltips with exact numbers of NA
values that are dropped per each column.
You may need to adjust some parameters before a histogram starts looking as you expect:
chart = na.altair.plot_hist(data, col='Pclass', col_na='Age')\
.properties(width=200, height=200)
chart.configure_axisX(labelAngle = 0)We can build a logistic regression model with Age as a dependent variable and
Fare, Parch, Pclass, SibSp, Survived as independent variables.
Internally, pandas.Series.isna() method is called on Age column, and the
resulting boolean values are converted to integers (True/False becomes
1/0). Finally, fitting a logistic model is done by
statsmodels package:
# Selecting columns with numeric data
# Dropping "PassengerId" column
subset = data.loc[:, data.dtypes != object].drop(columns=['PassengerId'])
model = na.model(subset, col_na='Age')
model.summary()Optimization terminated successfully.
Current function value: 0.467801
Iterations 7
Logit Regression Results
===============================================================================
Dep. Variable: Age No. Observations: 891
Model: Logit Df Residuals: 885
Method: MLE Df Model: 5
Date: Sat, 05 Jun 2021 Pseudo R-squ.: 0.06164
Time: 17:51:31 Log-Likelihood: -416.81
converged: True LL-Null: -444.19
Covariance Type: nonrobust LLR p-value: 1.463e-10
===============================================================================
coef std err z P>|z| [0.025 0.975]
-------------------------------------------------------------------------------
(intercept) -2.7294 0.429 -6.369 0.000 -3.569 -1.890
Fare 0.0010 0.003 0.376 0.707 -0.004 0.006
Parch -0.8874 0.223 -3.984 0.000 -1.324 -0.451
Pclass 0.5953 0.147 4.046 0.000 0.307 0.884
SibSp 0.2548 0.095 2.684 0.007 0.069 0.441
Survived -0.1026 0.198 -0.519 0.604 -0.490 0.285
===============================================================================
Use scikit_na.report() function to show interactive report interface:
na.report(data)Any contribution is highly appreciated: pull requests, suggestions, or bug reports.