Author: Javier Perales-Patón - @jperales
Programming at the cluster is not that straightforward since there is no graphical
interface. In contrast, an IDE fully based in the terminal is a powerful tool
for this task. In addition, important features of a good IDE includes a command-line
console to run lines of code, and auto-complete sentences as long as you are
programming.
Herein it is explained how to set-up such an IDE, so you could develop your project
directly when logged-in the cluster.
Unfortunately, terminal-based IDE are NOT friendly. It's highly recommended that
you have great expertise with the terminal, tmux (terminal client), and vim (plain text editor). Fortunately, there are good tutorials on the Internet
(e.g. vim tutorial).
What is this IDE about? You will have a terminal-based environment with the terminal split in two or three panels. First panel will allow you to program, second panel has an active command-line console session for the language interpreter (
R,python,bash, etc). Optionally, you can open this in atmuxpanel , so you keep a third panel withbashterminal forgit.
The program nvim is a requirement to be installed in the local/remote machine.
Then, you will need to install some plugins for nvim. To initiate the
config of nvim, create the following directory if it does not exist yet:
if[ ! -e $HOME/.config/nvim ];then
mkdir $HOME/.config/nvim
fi
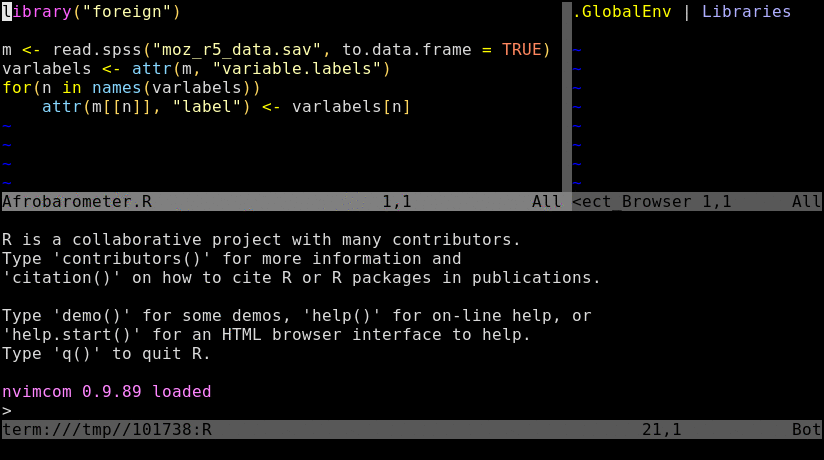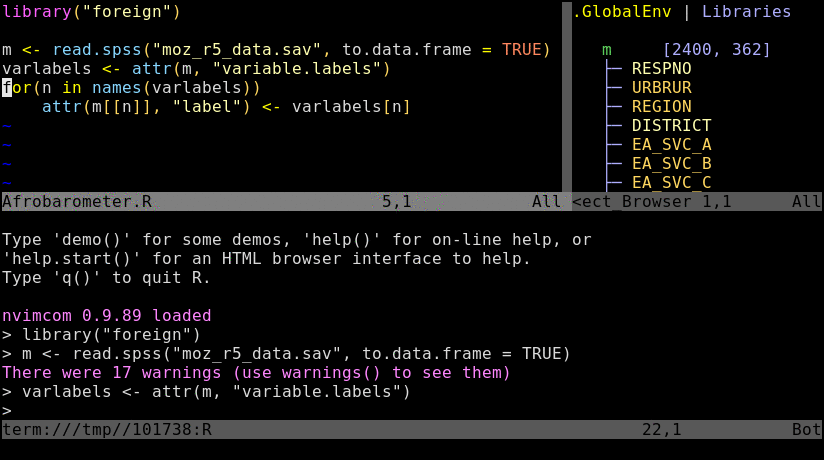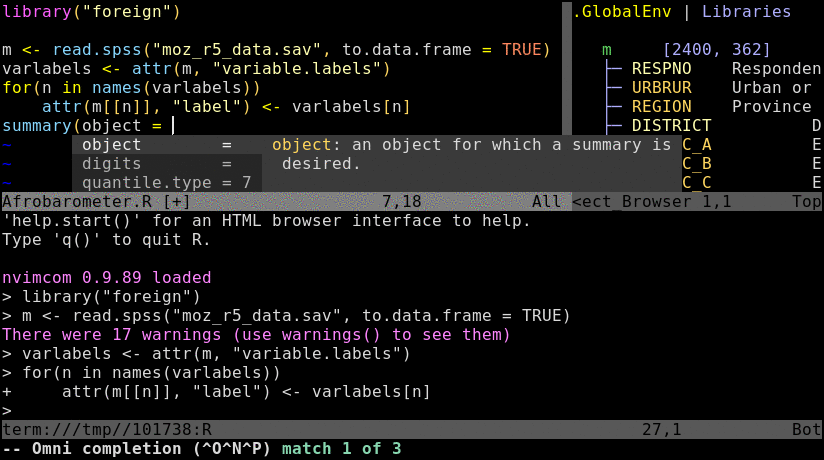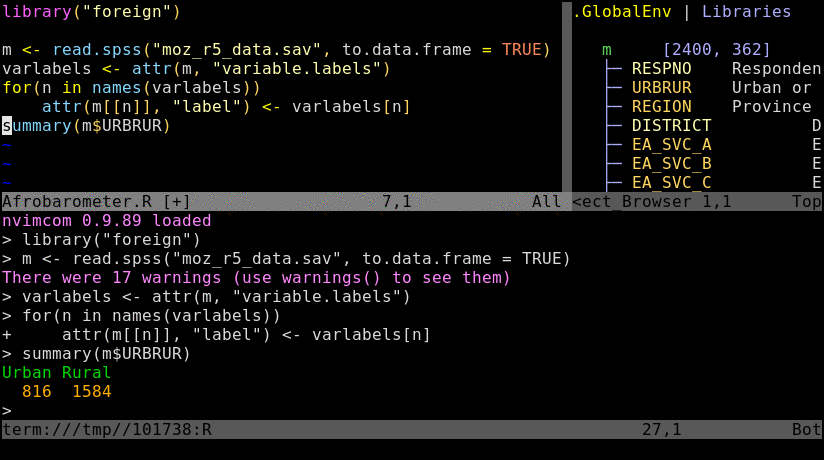
The nvim interface (first panel) communicates with the command-line interpreter allocated in a second panel.
To start the command-line interpreter for any language, you have to install certain ad-hoc plugins before.
You have to install vim-r. More info
Installation
Follow these instructions: https://gist.github.com/tgirke/7a7c197b443243937f68c422e5471899
Usage
Open a *.R or *.Rmd file with nvim and intialize a connected R session with \rf. This command can be remapped to other key combinations, e.g. uncommenting lines 10-12 in init.vim will remap it to the F2 key. Note, the resulting split window among Nvim and R behaves like a split viewport in nvim or vim meaning the usage of Ctrl-w w followed by i and Esc is important for navigation.
Important keybindings for nvim (vim):
\rf: opens vim-connected R sessionspacebar: sends code from vim to R; here remapped ininit.vimfrom default\l:splitor:vsplit: splits viewport (similar to pane split in tmux)gz: maximizes size of viewport in normal mode (similar to Tmux'sCtrl-a zzoom utility)Ctrl-w w: jumps cursor to R viewport and back; toggle between insert (i) and command (Esc) mode is required for navigation and controlling the environment.Ctrl-w r: swaps viewportsCtrl-w =: resizes splits to equal sizeCtrl-w 5< or 5>: resizes splits to left or right by 5 steps; change number as neededCtrl-w HorCtrl-w K: toggles between horizontal/vertical splitsCtrl-spacebar: omni completion for R objects/functions when nvim is in insert mode. Note, this has been remapped ininit.vimfrom difficult to type defaultCtrl-x Ctrl-o.:h nvim-R: opens nvim-R's user manual; navigation works the same as for any Vim/Nvim help document:Rhelp fct_name: opens help for a function from nvim's command mode with text completion supportCtrl-s and Ctrl-x: freezes/unfreezes vim (some systems)
You have to install vimcmdline. More info.
Installation
Copy the directories ftplugin, plugin and syntaxto your ~/.config/nvim
directory from this repository: https://github.com/jalvesaq/vimcmdline
Usage
If you are editing one of the supported file types, in Normal mode do:
\sto start the interpreter.<Space>to send the current line to the interpreter.\<Space>to send the current line to the interpreter and keep the cursor on the current line.\qto send the quit command to the interpreter. For languages that can source chunks of code:- In Visual mode, press:
<Space>to send a selection of text to the interpreter.
- And, in Normal mode, press:
\pto send from the line to the end of paragraph.\bto send block of code between the two closest marks.\fto send the entire file to the interpreter.
NOTE: official website https://cli.github.com Depending on Linux and user permisions, you could follow to alternatives:
- A) If root user, install it from official sources using apt-get install in Ubuntu, more details in https://github.com/cli/cli/blob/trunk/docs/install_linux.md#debian-ubuntu-linux-apt
sudo apt-key adv --keyserver keyserver.ubuntu.com --recv-key C99B11DEB97541F0
sudo apt-add-repository https://cli.github.com/packages
sudo apt update
sudo apt install gh
# If any error, follow the link above- B) If no root user, compile it for local user usage. It is far more complicated:
# Setting `go` intepreter for compilation
# Assuming you have a var in bash environment with such as,
# export SOFTWARE_LOCAL="/beegfs/work/$USER/SOFTWARE"
# 1. Download `go`
wget -O $SOFTWARE_LOCAL/go1.15.2.linux-amd64.tar.gz https://golang.org/dl/go1.15.2.linux-amd64.tar.gz
# SHA256 Checksum: b49fda1ca29a1946d6bb2a5a6982cf07ccd2aba849289508ee0f9918f6bb4552
# 2. Extract `go` as a local installation (src: https://golang.org/doc/install)
test -d $SOFTWARE_LOCAL/go1.15.2 || mkdir $SOFTWARE_LOCAL/go1.15.2
tar -C $SOFTWARE_LOCAL/go1.15.2 -xzf $SOFTWARE_LOCAL/go1.15.2.linux-amd64.tar.gz
# 3. Export it
# alias go='$SOFTWARE_LOCAL/go1.15.2/go/bin/go'
export PATH=$PATH:$SOFTWARE_LOCAL/go1.15.2/go/bin
# Test it
go -version
# Compile gh program from source
# 1. Clone repository
git clone https://github.com/cli/cli.git gh-cli
cd gh-cli
# 2. Build
make
# 3. Move the resulting bin/gh executable to somewhere in your PATH or create alias
# sudo mv ./bin/gh /usr/local/bin/
alias gh='$SOFTWARE_LOCAL/gh-cli/bin/gh'
# Test it
gh versionFinally, create a auth TOKEN following Settings> Developer settings > Personal access tokens.
Add the following permisions (at least, to be cross-checked with official doc):
- admin:org
- delete:packages
- delete_repo
- repo
- workflow
- write:packages
use gh auth login > choose Github.com & token auth. Paste it
A virtual environment (aka
env) is a self-contained local configuration of the basic software (compilers, interpreters, and packages) build-up for a specific use.
This is useful for:
- To circumvent outdated libraries from the cluster. For instance, the GCC compiler is outdated, which is required to compile third-party packages. You are going to face similar issues with JAVA (old v1.6), R (old v3.5). Using
envyou can use an updated compiler to complete the installation of your whole pipeline. - To use different versions of basic environments (R, python). For instance, some python programs require python 2.7, while other require python 3.0+. You may need to use both versions within a project.
- To keep independent configurations for different projects along time. One research project lasts for years and overlap along time with other ongoing projects.
envallows you to isolate software configurations across them, so updating packages for one project does not interfere with the settings from other projects. - Share your configuration with collaborators. You can clone your
envand share it with your colleagues, so they could reproduce your analysis.
miniconda3 is the framework to create and manage envs.
Before starting to work with virtual environments (envs), you need to install miniconda3. This is done only once.
Download the installer of miniconda3
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
Create a directory where you are going to install your SOFTWARE inside your working folder in the cluster
mkdir /home/$USER/SOFTWARE/
Run the installer and define the installation path. See below.
bash Miniconda3-latest-Linux-x86_64.sh
# Press <ENTER> to continue an read Miniconda License
# Go down and enter 'yes' to accept Miniconda License
# Importantly! Miniconda2 will be installed into the default location,
# that is your $HOME. However, there is no space in your $HOME at BioQuant.
# Thus, specify another location. For instance:
# /home/<username>/SOFTWARE/miniconda3
# run conda init when asked during the installation
If you didnt run conda init during the installation, this is a good time to do so. This will make changes into your ~/.bashrc file needed to setup the conda environment when needed.
Remove the installer
rm Miniconda3-latest-Linux-x86_64.sh
Basically you have to activate the env to work with, and deactivate it when you have finished your work.
Since the changes made in the ~/.bashrc file will take effect only after restart (which doesn't happen very often), to apply the changes you need to run
source ~./.bashrc
This will start a new bash environment with all profile and path settings needed for conda applied.
To activate an environment,
conda activate <env_name>
To deactivate it,
conda deactivate <env_name>
By default, you start with an env called base. You may give try with it.
You can also create new envs with your specific configuration.
Detailed documentation for Conda : further reading
Conda recipes are predefined list of packages with their corresponding dependencies ready to be installed at once in an existing or a new env.
There are several repositories (channels) throughby you can install these recipes. For instance, bioconda.
Some useful channels that you may add to your configuration are:
conda config --add channels conda-forge
conda config --add channels bioconda
conda config --add channels r
See the next section to understand how these channels can be useful.
Simply use
conda create -n <env_name> <recipe_1> <recipe_2> [...]
Where <env_name> is a short and unique name for the environment. And <recipe_N> is a recipe name from a channel.
Recommendation:
Use as many recipes as you can from the channels. These are shortcuts to install the package/software with all the dependencies. All bioconductor packages have a recipe...
How-To find recipes:
You can google
conda <package_name> recipe. Probably the 1st hit is a recipe from one of the channels (see Conda channels and recipes).
What if I cannot find a recipe for the package:
Then you have to install the
envwith the basics so far (e.g. R environment version 3.6, and other libraries and dependencies). Next install the package manually withenvactivated (see Installing software ). You can do this even without running an interactive session on the cluster.
You can install natively software at the same time that you create an env. This is very convenient. See Creating a virtual environment.
To install manually any package in the env, first you have to activate it
conda activate <env_name>
# See that the terminal prompt will show the <env_name>
# that means it is active
When an env is activated, some bash environment variables are updated/created:
$PATH- and other bash variables are updated. Thus the priority path to installed binary programs changes, so the binary ones from theenvare going to be used in preference to the others installed outside of theenv$CONDA_PREFIXis a new bash environment variable.$CONDA_PREFIXpoints out to the absolute to the local installation of the programs within theenv.
Note that this is an example for R packages, but similarly for other languages (python, java, etc).
- Activate the
env
conda activate <env_name>
- Start
R enviromentinside theenv:
$CONDA_PREFIX/bin/R
- Install your package as usual (inside
R). See Installing R packages for detailed information. You can do this even without running an interactive session on the cluster.
You have to consider two key points here:
- When you submit a job to the cluster, the cluster node starts a new session in UNIX with default settings.
- There are different binaries installed locally in the node. You may prefer to use one in particular. For that, you have to specify which one.
Thus you have include them in your PBS script :
- Activate the
envbefore running your script, adding the following line just after the header
conda activate <env_name>
Do not forget to add exec bash before this line to setup the conda environment, if the changes to the bash environment are still not automatically applied.
- [Optional] Specify the local program to run your script from the
envin order to avoid misinterpretations. For instance, for an R script:
# Use this one
$CONDA_PREFIX/bin/Rscript <your_script.R>
# ... Instead of
Rscript <your_script.R>
I am using Watson (poewred by tailor-dev team) at https://tailordev.github.io/Watson/ .
- Github: https://github.com/TailorDev/Watson
- User-Guide commands: https://tailordev.github.io/Watson/user-guide/commands/
How to install:
pip install td-watsonThen,
watson help