This repo is no longer maintained. For questions on this reop please email Honghan directly ([email protected]), or for broader CogStack enquiries please reach out to [email protected]
Surfacing Semantic Data from Clinical Notes in Electronic Health Records for Tailored Care, Trial Recruitment and Clinical Research
- Tutorial: use dockerised SemEHR on (dockerised) Elasticsearch (an example on 20 discharge summaries)
- Dockerised SemEHR
- Documentation on running SemEHR
- Documentation on API calls to SemEHR
- Minimised SemEHR + nlp2phenome bundle installation
- (02 Dec 2020) Minimised SemEHR / nlp2phenome bundle for restricted safe haven env (no Docker) has been created: https://github.com/CogStack/CogStack-SemEHR/tree/safehaven_mini/installation
- (03 June 2019) A tutorial to apply SemEHR on 20 discharge summaries and deploy the system on an Elasticsearch Cluster: Here
- (20 Dec 2018) Dead easy to run SemEHR NOW - Docker version of SemEHR released! Check docker.
- (26 Feb 2018) An actionable transparency model has been implemented to derive confidence/accuracy value for each annotation on a cohort basis. Such value is based on the syntactic/semantic/contextual characteristics of the containing sentence/document of annotations. (a working paper about this technique will be shared soon.)
- (9 Feb 2018) Patient Phenome UI implemented - to support 100k Genomics England (GeL) phenome model population for patients recruited for rare disease studies.
- (22 Dec 2017) An application paper describing SemEHR has been accepted by JAMIA, titled “SemEHR: A General-purpose Semantic Search System to Surface Semantic Data from Clinical Notes for Tailored Care, Trial Recruitment and Clinical Research”.
- (17 Nov 2017) Documentations for running SemEHR pipeline and API access have been put in the wiki: https://github.com/CogStack/SemEHR/wiki.
- (14 Sept 2017) An abstract describing SemEHR toolkit has been accepted to present at UK Publich Health Science Conference 2017 and to be published by The Lancet.
- (24 Apr 2017) An SemEHR instance has been deployed on MIMICIII data on 3 VMs of KCL's Rosalind HPC cluster to facilitate researches on this open ICU EHRs.
- (19 Apr 2017) The instance of SemEHR on SLaM's CRIS data is supportingt the discovery of associations between liver diseases and addictions.
- (21 Mar 2017) An SemEHR instance has been deployed at King's College Hospital to support patient recruitments for rare diseases in Genomics England's 100,000 Genome Project.
- (12 Oct 2016) SemEHR is initated as an effort to make use of EU KConnect results for supporting researches on anonymised EHR data of South London and Maudsley Hospital.
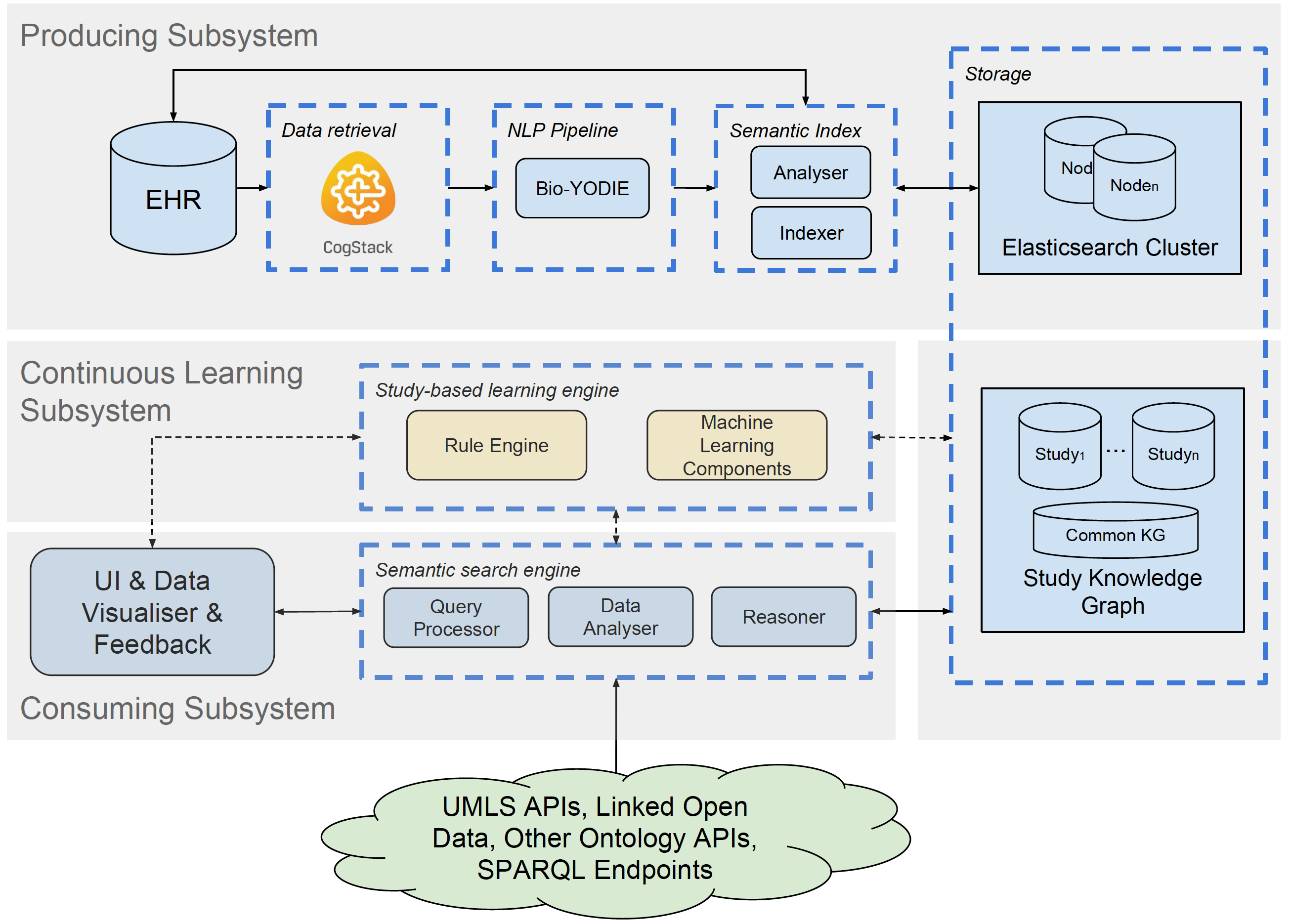
Built upon off-the-shelf toolkits including a Natural Language Processing (NLP) pipeline (Bio-Yodie) and an enterprise search system (CogStack), SemEHR implements a generic information extraction (IE) and retrieval infrastructure by identifying contextualised mentions of a wide range of biomedical concepts from unstructured clinical notes. Its IE functionality features an adaptive and iterative NLP mechanism where specific requirements and fine-tuning can be fulfilled and realised on a study basis. NLP annotations are further assembled at patient level and extended with clinical and EHR-specific knowledge to populate a panorama for each patient, which comprises a) longitudinal semantic data views and b) structured medical profile(s). The semantic data is serviced via ontology-based search and analytics interfaces to facilitate clinical studies.
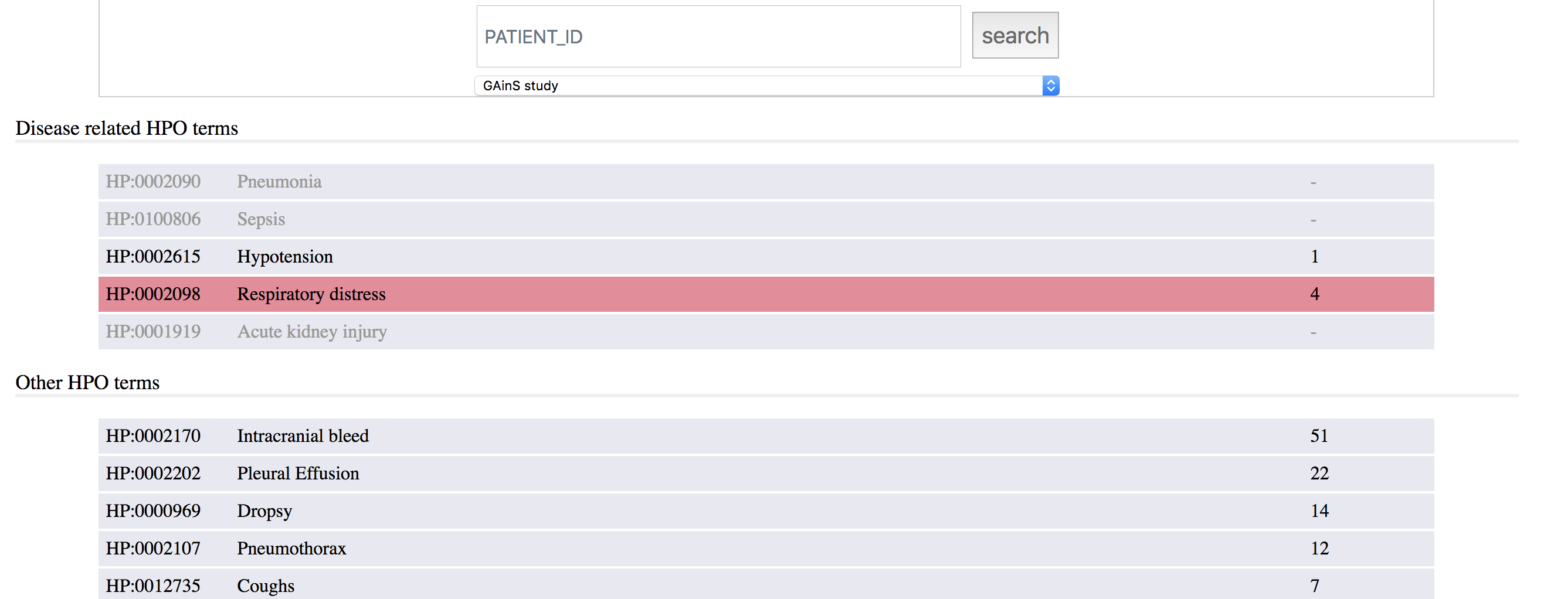
With SemEHR, the clinical variables hidden in clinical notes are surfaced via a set of search interfaces. A typical process to answer a clinical question (e.g. patients with hepatitis c), which previously might involve NLP, turns into one or a few google-style searches, for which SemEHR will pull out the cohort of relevant patients, populate patient-level summaries - numbers of contextualised concept mentions (e.g. 2nd patient has 16 total mentions of the disease, 15 of them were positive and 1 was historical), and link each mention to its original clinical note.
SemEHR: surfacing semantic data from clinical notes in electronic health records for tailored care, trial recruitment, and clinical research. Honghan Wu, Giulia Toti, Katherine I Morley, Zina Ibrahim, Amos Folarin, Ismail Kartoglu, Richard Jackson, Asha Agrawal, Clive Stringer, Darren Gale, Genevieve M Gorrell, Angus Roberts, Matthew Broadbent, Robert Stewart, Richard J B Dobson. The Lancet , Volume 390 , S97. 10.1016/S0140-6736(17)33032-5
SemEHR: A General-purpose Semantic Search System to Surface Semantic Data from Clinical Notes for Tailored Care, Trial Recruitment and Clinical Research. Honghan Wu, Giulia Toti, Katherine I Morley, Zina Ibrahim, Amos Folarin, Ismail Kartoglu, Richard Jackson, Asha Agrawal, Clive Stringer, Darren Gale, Genevieve M Gorrell, Angus Roberts, Matthew Broadbent, Robert Stewart, Richard J B Dobson.Journal of the American Medical Informatics Association, 2017. 10.1093/jamia/ocx160
Email Honghan Wu ([email protected])