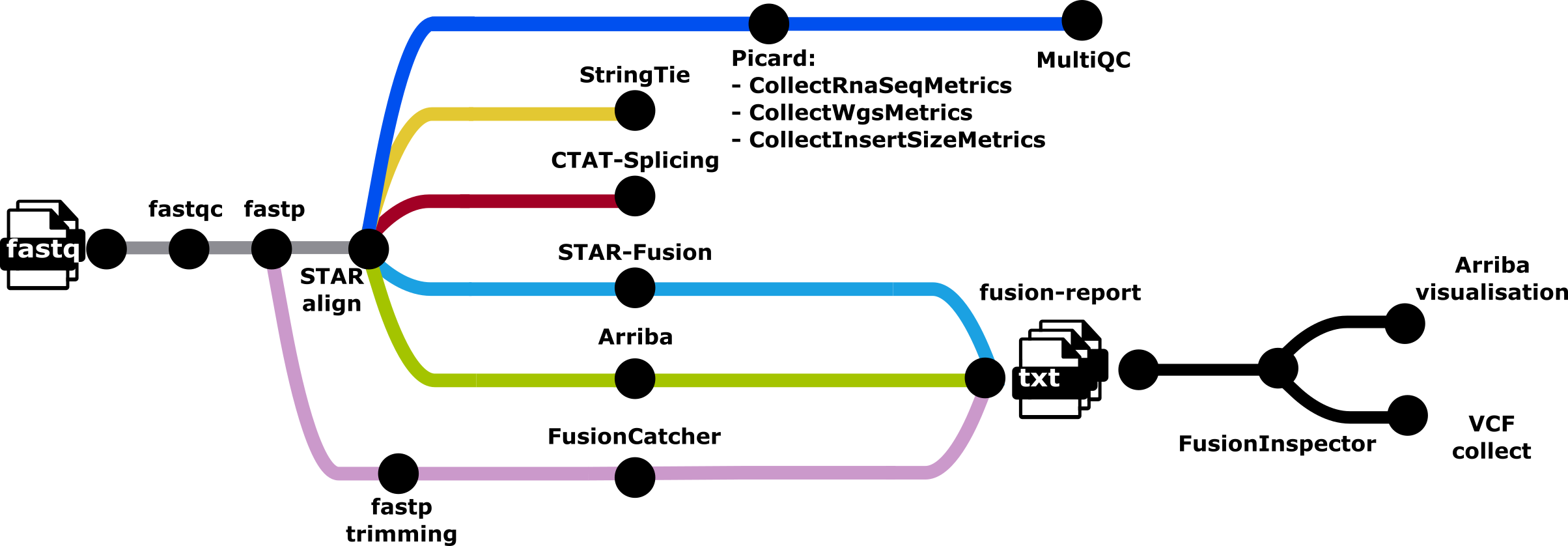
nf-core/rnafusion is a bioinformatics best-practice analysis pipeline for RNA sequencing analysis pipeline with curated list of tools for detecting and visualizing fusion genes.
The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It uses Docker/Singularity containers making installation trivial and results highly reproducible. The Nextflow DSL2 implementation of this pipeline uses one container per process which makes it much easier to maintain and update software dependencies. Where possible, these processes have been submitted to and installed from nf-core/modules in order to make them available to all nf-core pipelines, and to everyone within the Nextflow community!
IMPORTANT: conda is not supported currently. Run with singularity or docker.
GRCh38 is the only supported reference
| Tool | Single-end reads | Version |
|---|---|---|
| Arriba | ❌ | 2.3.0 |
| FusionCatcher | ✅ | 1.33 |
| Pizzly | ❌ | 0.37.3 |
| Squid | ❌ | 1.5 |
| STAR-Fusion | ✅ | 1.10.1 |
Single-end reads are to be use as last-resort. Paired-end reads are recommended. FusionCatcher cannot be used with single-end reads shorter than 130 bp.
On release, automated continuous integration tests run the pipeline on a full-sized dataset on the AWS cloud infrastructure. This ensures that the pipeline runs on AWS, has sensible resource allocation defaults set to run on real-world datasets, and permits the persistent storage of results to benchmark between pipeline releases and other analysis sources. The results obtained from the full-sized test can be viewed on the nf-core website.
In rnafusion the full-sized test includes reference building and fusion detection. The test dataset is taken from here.
--build_references triggers a parallel workflow to build all references
- Download ensembl fasta and gtf files
- Create STAR index
- Download arriba references
- Download fusioncatcher references
- Download pizzly references (kallisto index)
- Download and build STAR-fusion references
- Download fusion-report DBs
- Input samplesheet check
- Concatenate fastq files per sample
- Read QC (
FastQC) - Arriba subworkflow
- Pizzly subworkflow
- Squid subworkflow
- STAR alignment
- Samtools view: convert sam output from STAR to bam
- Samtools sort: bam output from STAR
- SQUID fusion detection
- SQUID annotate
- STAR-fusion subworkflow
- STAR alignment
- STAR-Fusion fusion detection
- Fusioncatcher subworkflow
- FusionCatcher fusion detection
- Fusion-report subworkflow
- Merge all fusions detected by the different tools
- Fusion-report
- FusionInspector subworkflow
- Present QC for raw reads (
MultiQC) - QC for mapped reads (
QualiMap: BAM QC) - Index mapped reads (samtools index)
- Collect metrics (
picard CollectRnaSeqMetricsand (picard MarkDuplicates)
-
Install
Nextflow(>=21.10.3) -
Install any of
Docker,Singularity(you can follow this tutorial),Podman,ShifterorCharliecloudfor full pipeline reproducibility (you can useCondaboth to install Nextflow itself and also to manage software within pipelines. Please only use it within pipelines as a last resort; see docs). -
Download the pipeline and test it on a minimal dataset with a single command:
nextflow run nf-core/rnafusion -profile test,YOURPROFILE --outdir <OUTDIR>Note that some form of configuration will be needed so that Nextflow knows how to fetch the required software. This is usually done in the form of a config profile (
YOURPROFILEin the example command above). You can chain multiple config profiles in a comma-separated string.- The pipeline comes with config profiles called
docker,singularity,podman,shifter,charliecloudandcondawhich instruct the pipeline to use the named tool for software management. For example,-profile test,docker. - Please check nf-core/configs to see if a custom config file to run nf-core pipelines already exists for your Institute. If so, you can simply use
-profile <institute>in your command. This will enable eitherdockerorsingularityand set the appropriate execution settings for your local compute environment. - If you are using
singularity, please use thenf-core downloadcommand to download images first, before running the pipeline. Setting theNXF_SINGULARITY_CACHEDIRorsingularity.cacheDirNextflow options enables you to store and re-use the images from a central location for future pipeline runs. - If you are using
conda, it is highly recommended to use theNXF_CONDA_CACHEDIRorconda.cacheDirsettings to store the environments in a central location for future pipeline runs.
- The pipeline comes with config profiles called
-
Start running your own analysis!
nextflow run nf-core/rnafusion --input samplesheet.csv --outdir <OUTDIR> --genome GRCh38 --all -profile <docker/singularity/podman/shifter/charliecloud/institute>Note that paths need to be absolute and that runs with conda are not supported.
The nf-core/rnafusion pipeline comes with documentation about the pipeline usage, parameters and output.
nf-core/rnafusion was written by Martin Proks (@matq007), Maxime Garcia (@maxulysse) and Annick Renevey (@rannick)
We thank the following people for their help in the development of this pipeline:
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don't hesitate to get in touch on the Slack #rnafusion channel (you can join with this invite).
If you use nf-core/rnafusion for your analysis, please cite it using the following doi: 10.5281/zenodo.3946477
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.