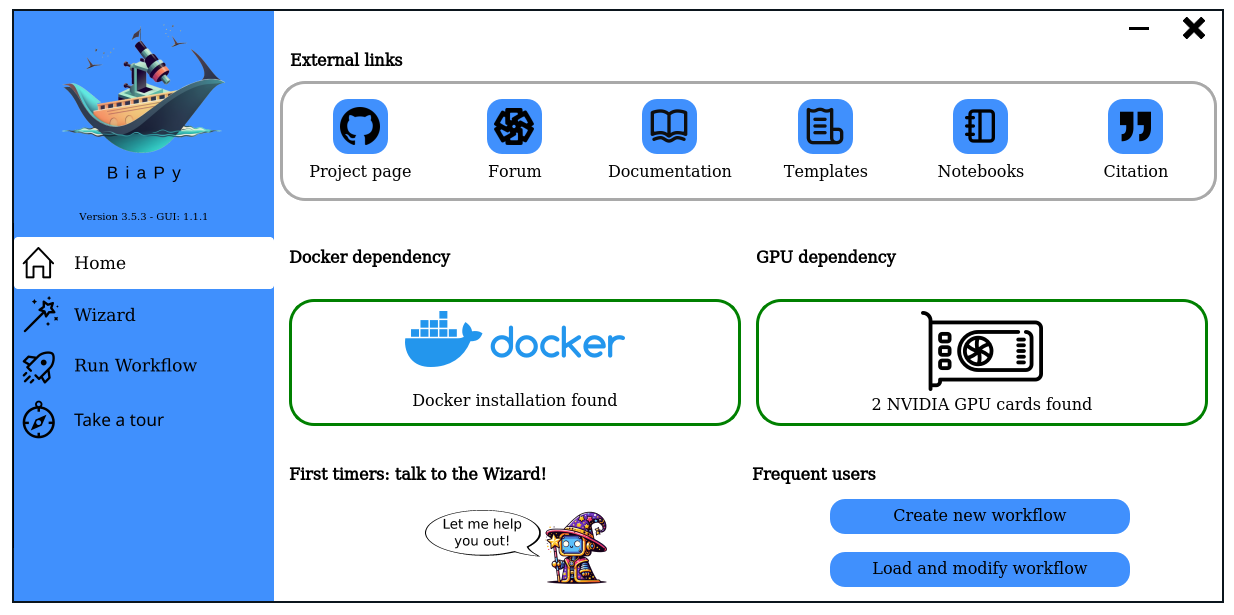
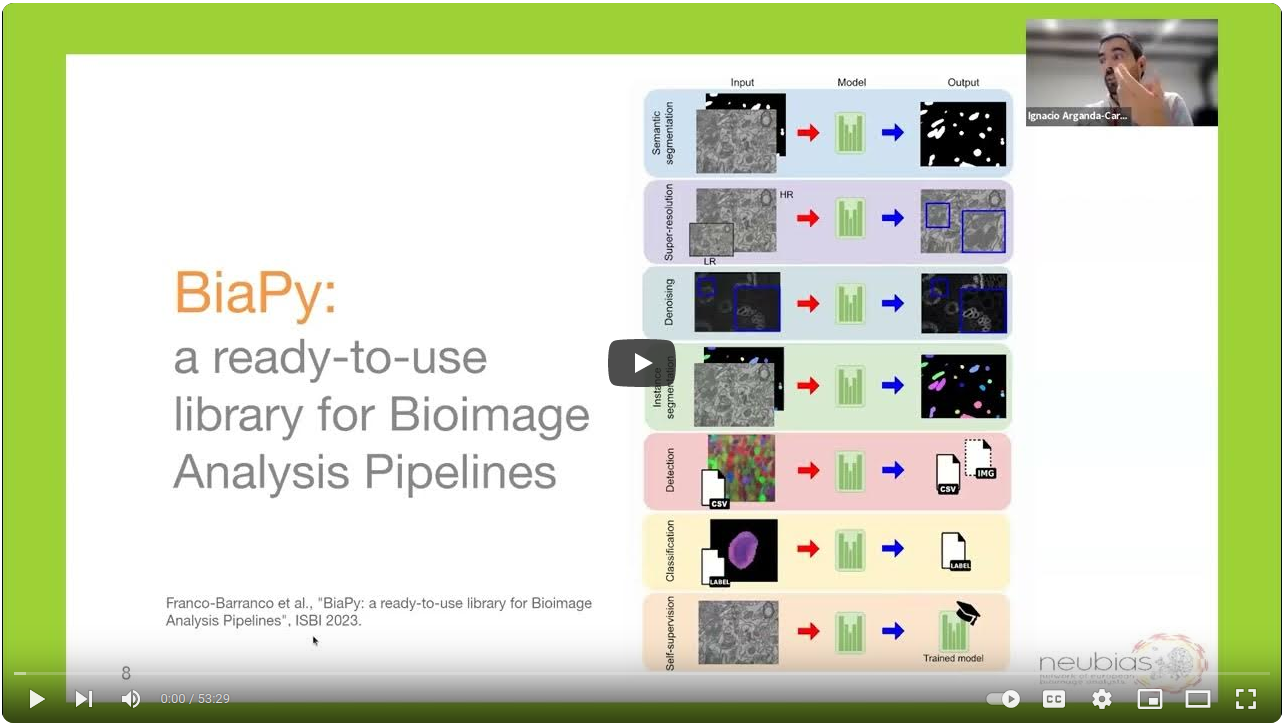
BiaPy is an open source ready-to-use all-in-one library that provides deep-learning workflows for a large variety of bioimage analysis tasks, including 2D and 3D semantic segmentation, instance segmentation, object detection, image denoising, single image super-resolution, self-supervised learning and image classification.
BiaPy is a versatile platform designed to accommodate both proficient computer scientists and users less experienced in programming. It offers diverse and user-friendly access points to our workflows.
This repository is actively under development by the Biomedical Computer Vision group at the University of the Basque Country and the Donostia International Physics Center.
Find a comprehensive overview of BiaPy and its functionality in the following videos:
 BiaPy history and GUI demo at RTmfm by Ignacio Arganda-Carreras and Daniel Franco-Barranco. |
|---|
 BiaPy presentation at Virtual Pub of Euro-BioImaging by Ignacio Arganda-Carreras. |
Download the repository first:
git clone https://github.com/BiaPyX/BiaPy-GUI.git
cd BiaPy-GUIThis will create a folder called BiaPy-GUI that contains all the files of the repository. Then you need to create a conda environment and install the dependencies:
conda create -n biapy-gui python=3.10.10
conda activate biapy-gui
pip install -r requirements.txtAfter that simply run the main.py :
python main.pyTo create the binary files:
pyinstaller -F main.pyYou can also modify the main.spec file (e.g. to add new images) and redo the binary:
pyinstaller main.specThis will create a main.spec file and a dist folder with BiaPy binary inside it.
Franco-Barranco, Daniel, et al. "BiaPy: a ready-to-use library for Bioimage Analysis Pipelines."
2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI). IEEE, 2023.