You signed in with another tab or window. Reload to refresh your session.You signed out in another tab or window. Reload to refresh your session.You switched accounts on another tab or window. Reload to refresh your session.Dismiss alert
This bug report is a reproducible example with code of an issue that may be in #7018, #2912 and other bug reports reporting slow performance and memory exhaustion when using .to_zarr() or .to_netcdf(). I think this has been hard to track down because it only occurs for large data sets. I have included code that replicates the problem without the need for downloading a large dataset.
The problem is that saving a xarray dataset which includes a variable with type datetime64[ns] is several orders of magnitude slower (!!) and uses a great deal of memory (!!) relative to the same dataset where that variable has another type. The work around is obvious -- turn off time decoding and treat time as a float64. But this is in-elegant, and I think this problem has lead to many un-answered questions on the issues page, such as the one above.
If I save a dataset whose structure (based on my use case, the ocean-parcels Lagrangian particle tracker) is:
<xarray.Dataset>
Dimensions: (trajectory: 953536, obs: 245)
Dimensions without coordinates: trajectory, obs
Data variables:
time (trajectory, obs) float32 dask.array<chunksize=(50000, 10), meta=np.ndarray>
age (trajectory, obs) float32 dask.array<chunksize=(50000, 10), meta=np.ndarray>
lat (trajectory, obs) float32 dask.array<chunksize=(50000, 10), meta=np.ndarray>
lon (trajectory, obs) float32 dask.array<chunksize=(50000, 10), meta=np.ndarray>
z (trajectory, obs) float32 dask.array<chunksize=(50000, 10), meta=np.ndarray>
I have no problems, even if the data set is much larger than the machine's memory. However, if I change the time variable to have the data type datetime64[ns]
<xarray.Dataset>
Dimensions: (trajectory: 953536, obs: 245)
Dimensions without coordinates: trajectory, obs
Data variables:
time (trajectory, obs) datetime64[ns] dask.array<chunksize=(50000, 10), meta=np.ndarray>
age (trajectory, obs) float32 dask.array<chunksize=(50000, 10), meta=np.ndarray>
lat (trajectory, obs) float32 dask.array<chunksize=(50000, 10), meta=np.ndarray>
lon (trajectory, obs) float32 dask.array<chunksize=(50000, 10), meta=np.ndarray>
z (trajectory, obs) float32 dask.array<chunksize=(50000, 10), meta=np.ndarray>
the time it takes to write this dataSet becomes much greater, and increases much more quickly with an increase in the "trajectory" coordinate then the case where time has type "float64." The increase in time is NOT in the writing time, but in the time it takes to compute the dask graph. At the same time the time to compute the graph increases, the memory usage increases, finally leading to memory exhaustion as the data set gets larger. This can be seen in the attached figure, which shows the time to create the graph with dataOut.to_zarr(outputDir,compute=False) and the time to write the data with delayedObj.compute(). By the time the data set is 10 million long in the first dimension, the dataset with datetime64[ns] takes 4 orders of magnitude longer (!!!) to compute -- hours instead of seconds!
To recreate this graph, and to see a very simple code that replicates this problem, see the attached python code. Note that the directory you run it in should have at least 30Gb free for the data set it writes, and for machines with less than 256Gb of memory, it will crash before completing after exhausting the memory. However, the last figure will be saved in jnk_out.png, and you can always change the largest size it attempts to create.
I expect that the time to save a dataset with .to_zarr or .to_netcdf does not change dramatically if one of the variables has a datetime64[ns] type.
Minimal Complete Verifiable Example
#this code is also included as a zip file above. importxarrayasxrfrompylabimport*fromnumpyimport*#from glob import glob#from os import pathimporttimeimportdaskfromdask.diagnosticsimportProgressBarimportshutilimportpickle#this is a minimal code that illustrates issue with .to_zarr() or .to_netcdf when writing a dataset with datetime64 data#outputDir is the name of the zarr output; it should be set to a location on a fast filesystem with enough spaceoutputDir='./testOut.zarr'deftestToZarr(dimensions,haveTimeType=True):
'''This code writes out an empty dataset with the dimensions specified in the "dimensions" arguement, and returns the time it took to create the dask delayed object and the time it took to compute the delayed object. if haveTimeType is True, the "time" variable has type datetime64 '''ifhaveTimeType:
#specify the type of variables to be written out. Each has dimensions (trajectory,obs)varType={'time':dtype('datetime64[ns]'),
'age':dtype('float32'),
'lat':dtype('float32'),
'lon':dtype('float32'),
'z':dtype('float32'),
}
else:
varType={'time':dtype('float32'),
'age':dtype('float32'),
'lat':dtype('float32'),
'lon':dtype('float32'),
'z':dtype('float32'),
}
#now make an empty dataset dataOut=xr.Dataset()
#now add the empty variablesforvinvarType:
vEmpty=dask.array.zeros((dimensions['trajectory'],dimensions['obs']),dtype=varType[v])
dataOut=dataOut.assign({v:(('trajectory','obs'),vEmpty)})
#chunk datachunksize={'trajectory':5*int(1e4),'obs':10}
print(' chunking dataset to',chunksize)
dataOut=dataOut.chunk(chunksize)
#create dask delayed object, and time how long it tooktic=time.time()
ifTrue: #write to zarrdelayedObj=dataOut.to_zarr(outputDir,compute=False)
else: #write to netCDFdelayedObj=dataOut.to_netcdf(outputDir,compute=False)
createGraphTime=time.time()-ticprint(' created graph in',createGraphTime)
#execute the delayed object, and see how long it took. Use progress bartic=time.time()
withProgressBar():
results=delayedObj.compute()
writeOutTime=time.time()-ticprint(' wrote data in',writeOutTime)
returncreateGraphTime,writeOutTime,dataOut#now lets do some benchmarkingif__name__=="__main__":
figure(1,figsize=(10.0,8.0))
clf()
style.use('ggplot')
#make a vector that is the size of the first dimension#("trajectory") of the data settrajectoryVec=logspace(log10(1000),log10(1.4e8),20).astype(int)
#pre-allocate variables to store resultscreateGraphTimeVec_time=0.0*trajectoryVec+nanwriteOutTimeVec_time=0.0*trajectoryVec+nancreateGraphTimeVec_noTime=0.0*trajectoryVec+nanwriteOutTimeVec_noTime=0.0*trajectoryVec+nan#get data for various array sizesforninrange(len(trajectoryVec)):
#write out data, and benchmark timedimensions={'trajectory':trajectoryVec[n],'obs':245}
print('starting to write file of dimensions',dimensions,'for case with time variable')
shutil.rmtree(outputDir,ignore_errors=True)
createGraphTimeVec_time[n],writeOutTimeVec_time[n],dataOut_time=testToZarr(dimensions,haveTimeType=True)
shutil.rmtree(outputDir,ignore_errors=True)
createGraphTimeVec_noTime[n],writeOutTimeVec_noTime[n],dataOut_noTime=testToZarr(dimensions,haveTimeType=False)
print(' done')
#now plot outputclf()
subplot(2,1,1)
loglog(trajectoryVec,createGraphTimeVec_time,'r-o',label='with datetime64 variable')
loglog(trajectoryVec,createGraphTimeVec_noTime,'b-*',label='without datetime64 variable')
#xlabel('size of first dimensions')ylabel('seconds')
title('time to create dask graph',fontsize='medium')
legend()
firstAx=axis()
subplot(2,1,2)
loglog(trajectoryVec,writeOutTimeVec_time,'r-o',label='with datetime64 variable')
loglog(trajectoryVec,writeOutTimeVec_noTime,'b-*',label='without datetime64 variable')
axis(xmin=firstAx[0],xmax=firstAx[1])
xlabel('size of first dimensions')
ylabel('seconds')
title('time to write data to disk',fontsize='medium')
legend()
draw()
show()
pause(0.01)
#save the figure each time, since this code can crash as the#size of the dataset gets larger and the dataset with#datetime64[ns] causes .to_zarr() to exhaust all the memorysavefig('jnk_out.png',dpi=100)
MVCE confirmation
Minimal example — the example is as focused as reasonably possible to demonstrate the underlying issue in xarray.
Complete example — the example is self-contained, including all data and the text of any traceback.
Verifiable example — the example copy & pastes into an IPython prompt or Binder notebook, returning the result.
New issue — a search of GitHub Issues suggests this is not a duplicate.
Relevant log output
No response
Anything else we need to know?
No response
Environment
Note -- I see the same thing on my linux machine
INSTALLED VERSIONS
------------------
commit: None
python: 3.10.6 | packaged by conda-forge | (main, Aug 22 2022, 20:41:22) [Clang 13.0.1 ]
python-bits: 64
OS: Darwin
OS-release: 21.6.0
machine: arm64
processor: arm
byteorder: little
LC_ALL: None
LANG: en_US.UTF-8
LOCALE: ('en_US', 'UTF-8')
libhdf5: None
libnetcdf: None
JamiePringle
changed the title
.to_zarr() or .to_netcdf slow and uses memory when datetime64[ns] variable in output; a reproducible example
.to_zarr() or .to_netcdf slow and uses excess memory when datetime64[ns] variable in output; a reproducible example
Sep 13, 2022
What happened?
This bug report is a reproducible example with code of an issue that may be in #7018, #2912 and other bug reports reporting slow performance and memory exhaustion when using .to_zarr() or .to_netcdf(). I think this has been hard to track down because it only occurs for large data sets. I have included code that replicates the problem without the need for downloading a large dataset.
The problem is that saving a xarray dataset which includes a variable with type datetime64[ns] is several orders of magnitude slower (!!) and uses a great deal of memory (!!) relative to the same dataset where that variable has another type. The work around is obvious -- turn off time decoding and treat time as a float64. But this is in-elegant, and I think this problem has lead to many un-answered questions on the issues page, such as the one above.
If I save a dataset whose structure (based on my use case, the ocean-parcels Lagrangian particle tracker) is:
I have no problems, even if the data set is much larger than the machine's memory. However, if I change the time variable to have the data type datetime64[ns]
the time it takes to write this dataSet becomes much greater, and increases much more quickly with an increase in the "trajectory" coordinate then the case where time has type "float64." The increase in time is NOT in the writing time, but in the time it takes to compute the dask graph. At the same time the time to compute the graph increases, the memory usage increases, finally leading to memory exhaustion as the data set gets larger. This can be seen in the attached figure, which shows the time to create the graph with
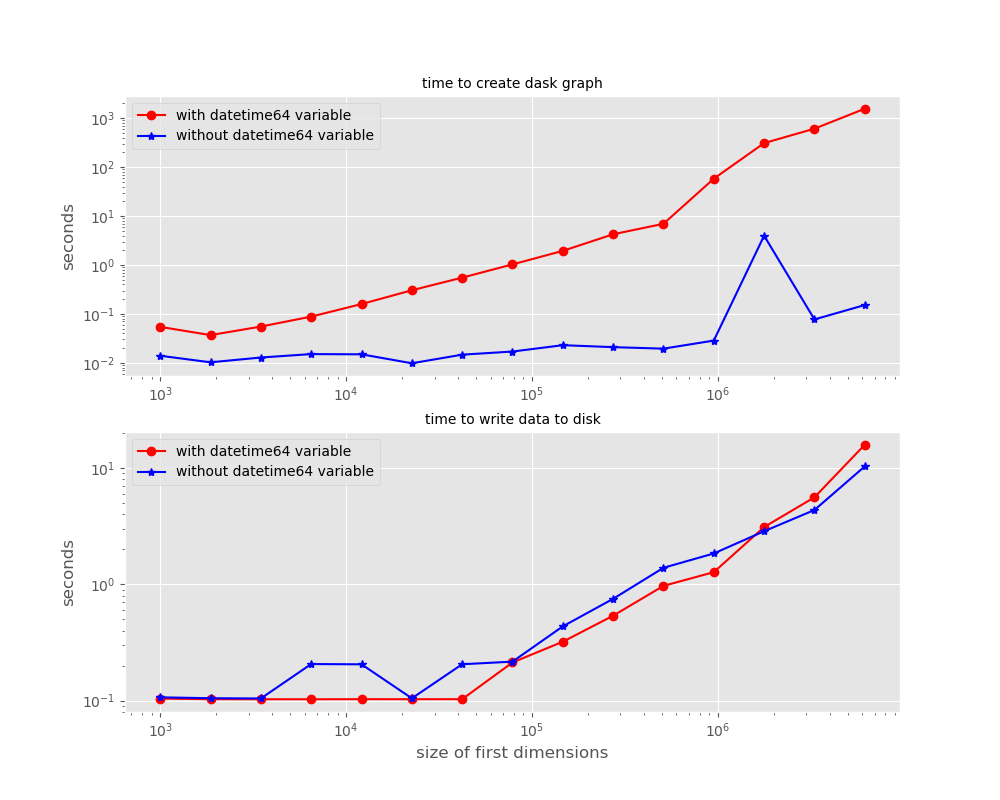
dataOut.to_zarr(outputDir,compute=False)and the time to write the data withdelayedObj.compute(). By the time the data set is 10 million long in the first dimension, the dataset with datetime64[ns] takes 4 orders of magnitude longer (!!!) to compute -- hours instead of seconds!To recreate this graph, and to see a very simple code that replicates this problem, see the attached python code. Note that the directory you run it in should have at least 30Gb free for the data set it writes, and for machines with less than 256Gb of memory, it will crash before completing after exhausting the memory. However, the last figure will be saved in jnk_out.png, and you can always change the largest size it attempts to create.
SmallestExample_zarrOutProblem.zip
What did you expect to happen?
I expect that the time to save a dataset with .to_zarr or .to_netcdf does not change dramatically if one of the variables has a datetime64[ns] type.
Minimal Complete Verifiable Example
MVCE confirmation
Relevant log output
No response
Anything else we need to know?
No response
Environment
Note -- I see the same thing on my linux machine
xarray: 2022.6.0
pandas: 1.4.3
numpy: 1.23.2
scipy: 1.9.0
netCDF4: None
pydap: None
h5netcdf: None
h5py: None
Nio: None
zarr: 2.12.0
cftime: None
nc_time_axis: None
PseudoNetCDF: None
rasterio: None
cfgrib: None
iris: None
bottleneck: None
dask: 2022.8.1
distributed: 2022.8.1
matplotlib: 3.5.3
cartopy: 0.20.3
seaborn: None
numbagg: None
fsspec: 2022.7.1
cupy: None
pint: None
sparse: None
flox: None
numpy_groupies: None
setuptools: 65.3.0
pip: 22.2.2
conda: None
pytest: None
IPython: 8.4.0
sphinx: None
The text was updated successfully, but these errors were encountered: