- pre-processing (motion correction, spatiotemporal downsampling, spatial filtering, relative fluorescence calculation, etc.) - support for multiple cell-extraction methods (CELLMax, PCA-ICA, CNMF, CNMF-E, EXTRACT, etc.) - manual classification of cells via GUIs, - - automated cell classification (coming soon!), + - automated cell classification (i.e. CLEAN algorithm, coming soon!), - cross-session cell alignment, and more. - Includes example one- and two-photon calcium imaging datasets for testing `ciatah`. -- Animal position tracking (e.g. in open-field assay). -- Supports most major imaging movie file formats: HDF5, NWB, AVI, ISXD [Inscopix], and TIFF. +- Supports a plethora of major imaging movie file formats: HDF5, NWB, AVI, ISXD [Inscopix], TIFF, and [Bio-Formats](https://www.openmicroscopy.org/bio-formats/) compatible formats (Olympus [OIR] and Zeiss [CZI and LSM] currently, additional support to be added or upon request). - Supports [Neurodata Without Borders](https://www.nwb.org/) data standard (see [calcium imaging tutorial](https://neurodatawithoutborders.github.io/matnwb/tutorials/html/ophys.html)) for reading/writing cell-extraction and imaging movie files. +- Animal position tracking (e.g. in open-field assay). - Requires `MATLAB`. @@ -114,7 +121,7 @@ Made in USA.
Below are steps needed to quickly get started using the `CIAtah` software package in MATLAB. -### Install +### Download and install `CIAtah` - Clone the `CIAtah` repository (using [GitHub desktop](https://desktop.github.com/) or command line) or download the repository zip and unzip (e.g. run below MATLAB command). - Point the MATLAB path to the `CIAtah` root folder (*NOT* `@CIAtah` sub-folder in the repository). @@ -132,6 +139,14 @@ Below are steps needed to quickly get started using the `CIAtah` software packag cd('ciatah-master') ``` +### Check required toolboxes are installed + +`CIAtah` depends on several MATLAB toolboxes to run properly. Run the below command to have `CIAtah` check whether dependencies have been installed properly. If not use the `Add-Ons` (https://www.mathworks.com/help/matlab/matlab_env/get-add-ons.html) explorer to install each toolbox. + +```Matlab +ciapkg.io.matlabToolboxCheck;` +``` + ### Setup `CIAtah` - Run `CIAtah` using the below MATLAB commands. Call `obj;` in the MATLAB command window each time you want to go back to the main GUI. __Note: `calciumImagingAnalysis` class is now called `ciatah`, all functionality is the same.__ @@ -151,6 +166,7 @@ obj % then hit enter, no semicolon! - Run `obj;` in the command window to see the main GUI. - Full documentation at https://bahanonu.github.io/ciatah/. - __[Optional]__ Users on Windows systems can download `Everything` (https://www.voidtools.com/). It is a very useful and extremely fast search engine for files and folders that allows users to quickly obtain lists of full folder paths for analysis in `CIAtah`. +- __[Optional]__ If run into issues opening certain AVI files (e.g. due to codec issues) with CIAtah/MATLAB, install `K-Lite Codec Pack` (https://codecguide.com/download_kl.htm) or similar for added support. ### Visualize movies quickly using read from disk diff --git a/ciapkg/classification/signalSorter.m b/ciapkg/classification/signalSorter.m index 7b5fb16..74ea6af 100644 --- a/ciapkg/classification/signalSorter.m +++ b/ciapkg/classification/signalSorter.m @@ -71,6 +71,7 @@ % 2020.10.13 [01:22:47] - Display context menu for keyboard shortcuts, easier than separate figure. User can select with right-click or via a menu in the GUI. % 2020.10.13 [22:31:23] - Users can now scroll through cells using mouse scroll wheel. % 2021.03.10 [16:33:23] - User can now input just NWB path without a blank variable for inputSignals. Also added support for CIAtah mat files. + % 2021.05.09 [15:20:18] - Since ciapkg.io.loadSignalExtraction already supports loading NWB or CIAtah MAT-file cell extraction files, remove remove redundancy and only call ciapkg.io.loadSignalExtraction rather than loadNeurodataWithoutBorders. Added support for progress bar when movie not input. % TODO % DONE: New GUI interface to allow users to scroll through video and see cell activity at that point % DONE: allow option to mark rest as bad signals @@ -244,17 +245,8 @@ % If inputs are NWB or CIAtah mat format if nargin==1|ischar(inputImages) - [~,~,inputEXT] = fileparts(inputImages); - switch inputEXT - case '.nwb' - [inputImages,inputSignals,infoStruct,algorithmStr] = loadNeurodataWithoutBorders(inputImages); - options.nSignals = size(inputImages,3); - case '.mat' - [inputImages,inputSignals,infoStruct,algorithmStr] = ciapkg.io.loadSignalExtraction(inputImages); - options.nSignals = size(inputImages,3); - otherwise - - end + [inputImages,inputSignals,infoStruct,algorithmStr] = ciapkg.io.loadSignalExtraction(inputImages); + options.nSignals = size(inputImages,3); end % Modify dataset name if given NWB file @@ -396,6 +388,7 @@ options.movieMin = 0; end + % ===================== % Set all starting values to neutral (e.g. gray in GUI, to reduce bias). if strcmp(valid,'neutralStart')==1 valid = zeros([options.nSignals 1],'logical')+3; @@ -797,6 +790,9 @@ function plotSignalStatistics(inputSignals,inputImageSizes,inputStr,pointColor, % loop over chosen filters nImages = size(inputImages,3); + axValid = []; + axValidAll = []; + % initialize loop variables saveData=0; i = 1; % the current signal # being sorted @@ -1022,8 +1018,10 @@ function plotSignalStatistics(inputSignals,inputImageSizes,inputStr,pointColor, axis equal tight; end s2Pos = plotboxpos(inputMoviePlotLoc2Handle); - cbh = colorbar(inputMoviePlotLoc2Handle,'Location','eastoutside','Position',[s2Pos(1)+s2Pos(3)+0.005 s2Pos(2) 0.01 s2Pos(4)],'FontSize',options.fontSize-2); - ylabel(cbh,'Fluorescence (e.g. \DeltaF/F or \DeltaF/\sigma)','FontSize',options.fontSize-1); + if loopCount==1 + cbh = colorbar(inputMoviePlotLoc2Handle,'Location','eastoutside','Position',[s2Pos(1)+s2Pos(3)+0.005 s2Pos(2) 0.01 s2Pos(4)],'FontSize',options.fontSize-2); + ylabel(cbh,'Fluorescence (e.g. \DeltaF/F or \DeltaF/\sigma)','FontSize',options.fontSize-1); + end end catch err @@ -1056,11 +1054,12 @@ function plotSignalStatistics(inputSignals,inputImageSizes,inputStr,pointColor, else set(mainFig,'CurrentAxes',inputMoviePlotLoc2Handle); end - [thisImageCrop] = subfxnCropImages(thisImage); - imagesc(thisImageCrop); - % colormap gray - axis off; % ij square - title(['signal ' cellIDStr 10 '(' num2str(sum(valid==1)) ' good)'],'FontSize',options.fontSize,'Interpreter','tex'); + + [thisImageCrop] = subfxnCropImages(thisImage); + imagesc(thisImageCrop); + % colormap gray + axis off; % ij square + title(['signal ' cellIDStr 10 '(' num2str(sum(valid==1)) ' good)'],'FontSize',options.fontSize,'Interpreter','tex'); end % use thresholded image as AlphaData to overlay on cell map, reduce number of times this is accessed to speed-up analysis @@ -1368,6 +1367,7 @@ function plotSignalStatistics(inputSignals,inputImageSizes,inputStr,pointColor, set(gcf,'pointer','custom','PointerShapeCData',NaN([16 16])); end + % Display options if movie present if ~isempty(options.inputMovie) try if options.peakTrigMovieMontage==0 @@ -1513,14 +1513,6 @@ function plotSignalStatistics(inputSignals,inputImageSizes,inputStr,pointColor, title(['Press Q to change movie contrast.' 10 'Press Z to turn zoom on.' 10 'Press B to switch to movie montage.' 10 'Zoom enabled in trace panel.'],'FontSize',options.fontSize,'Interpreter','tex') end - % ========================================== - % ADD PROGRESS BAR - if exist('axValid','var')==1 - [axValid axValidAll] = subfxnSignalSorterProgressBars(i,valid,inputMoviePlotLocHandle,inputMoviePlotLoc2Handle,options,mainFig,axValid,axValidAll,cellIDStr); - else - [axValid axValidAll] = subfxnSignalSorterProgressBars(i,valid,inputMoviePlotLocHandle,inputMoviePlotLoc2Handle,options,mainFig,[],[],cellIDStr); - end - set(mainFig,'CurrentAxes',inputMoviePlotLocHandle); frameNoTotal = 0; % use this to lock @@ -1541,6 +1533,9 @@ function plotSignalStatistics(inputSignals,inputImageSizes,inputStr,pointColor, set(objMapZoomPlotLocHandle,'ButtonDownFcn',@subfxnSelectCellOnCellmap) set(mainFig, 'KeyPressFcn', @(source,eventdata) figure(mainFig)); + % Create progress bar + [axValid, axValidAll] = subfxn_progressBarCreate(axValid, axValidAll); + while strcmp(keyIn,'3') frameNoTotal = frameNoTotal+1; if frameNoTotal>3 @@ -1576,9 +1571,17 @@ function plotSignalStatistics(inputSignals,inputImageSizes,inputStr,pointColor, end else keyIn = get(gcf,'CurrentCharacter'); + % Create progress bar + [axValid, axValidAll] = subfxn_progressBarCreate(axValid, axValidAll); + + while strcmp(keyIn,'3') + keyIn = get(gcf,'CurrentCharacter'); + pause(1/options.fps); + end reply = double(keyIn); set(gcf,'currentch','3'); end + figure(mainFig); warning('on','all') @@ -1609,7 +1612,7 @@ function plotSignalStatistics(inputSignals,inputImageSizes,inputStr,pointColor, end end % loop if user gets to either end - i=i+directionOfNextChoice; + i = i+directionOfNextChoice; if i<=0; i = nImages; end if i>nImages; i = 1; end figure(mainFig); @@ -1629,6 +1632,26 @@ function plotSignalStatistics(inputSignals,inputImageSizes,inputStr,pointColor, warning('on','all') warning('query','all') safeExit = 1; + + function [axValid, axValidAll] = subfxn_progressBarCreate(axValid, axValidAll) + % ========================================== + % ADD PROGRESS BAR + if isempty(options.inputMovie) + tmpHandleProgressBar = avgSpikeTracePlotHandle; + tmpHandleProgressBar2 = inputMoviePlotLoc2Handle; + else + tmpHandleProgressBar = inputMoviePlotLocHandle; + tmpHandleProgressBar2 = inputMoviePlotLoc2Handle; + end + if isempty(axValid)==1 + % if exist('axValid','var')==0 + % if i==1&loopCount==1 + disp('Creating sorter progress bar...') + [axValid, axValidAll] = subfxnSignalSorterProgressBars(i,valid,tmpHandleProgressBar,tmpHandleProgressBar2,options,mainFig,[],[],cellIDStr); + else + [axValid, axValidAll] = subfxnSignalSorterProgressBars(i,valid,tmpHandleProgressBar,tmpHandleProgressBar2,options,mainFig,axValid,axValidAll,cellIDStr); + end + end function subfxnUserInputGui(replyTmp) if nargin>0 % Change keyIn to force while loop to exit, need for certain commands. @@ -2184,7 +2207,7 @@ function movieCallback(source,eventdata) end end end -function [axValid axValidAll] = subfxnSignalSorterProgressBars(i,valid,inputMoviePlotLocHandle,inputMoviePlotLoc2Handle,options,mainFig,axValid,axValidAll,cellIDStr) +function [axValid, axValidAll] = subfxnSignalSorterProgressBars(i,valid,inputMoviePlotLocHandle,inputMoviePlotLoc2Handle,options,mainFig,axValid,axValidAll,cellIDStr) validData = cat(3,... double(valid(:)'==0|valid(:)'>1),...% Red double(valid(:)'==1|valid(:)'>1),...% Green @@ -2206,6 +2229,7 @@ function movieCallback(source,eventdata) cbh = colorbar(inputMoviePlotLoc2Handle,'Location','eastoutside','Position',[s2Pos(1)+s2Pos(3)+0.005 s2Pos(2) 0.01 s2Pos(4)],'FontSize',options.fontSize-2); ylabel(cbh,'Fluorescence (e.g. \DeltaF/F or \DeltaF/\sigma)','FontSize',options.fontSize-1); + set(mainFig,'CurrentAxes',inputMoviePlotLocHandle); s3Pos = plotboxpos(gca); axValid = axes('Position',[s3Pos(1) 0.98 s3Pos(3) 0.02],'XTick',[],'YTick',[]); diff --git a/ciapkg/download/downloadGithubRepositories.m b/ciapkg/download/downloadGithubRepositories.m index 6c18ca0..a79de9c 100644 --- a/ciapkg/download/downloadGithubRepositories.m +++ b/ciapkg/download/downloadGithubRepositories.m @@ -104,7 +104,13 @@ % Rename to proper folder for calciumImagingAnalysis fprintf('Renaming %s to %s \n',[signalExtractionDir filesep gitName{gitNo}],[signalExtractionDir filesep outputDir{gitNo}]) - movefile([signalExtractionDir filesep gitName{gitNo}],[signalExtractionDir filesep outputDir{gitNo}]); + oldDir = [signalExtractionDir filesep gitName{gitNo}]; + newDir = [signalExtractionDir filesep outputDir{gitNo}]; + if strcmp(oldDir,newDir)==1 + disp('Same directory, ignore name change!') + else + movefile(oldDir,newDir); + end % fprintf('\n\n') end diff --git a/ciapkg/image/cropMatrix.m b/ciapkg/image/cropMatrix.m index d4165fe..84aff21 100644 --- a/ciapkg/image/cropMatrix.m +++ b/ciapkg/image/cropMatrix.m @@ -1,4 +1,4 @@ -function [inputMatrix] = cropMatrix(inputMatrix,varargin) +function [inputMatrix, coords] = cropMatrix(inputMatrix,varargin) % Crops a matrix either by removing rows or adding NaNs to where data was previously. % Biafra Ahanonu % 2014.01.23 [16:06:01] @@ -7,7 +7,8 @@ % outputs % inputMatrix - cropped or NaN'd matrix, same name to reduce memory usage % changelog - % 2017.01.14 [20:06:04] - support switched from [nSignals x y] to [x y nSignals] + % 2017.01.14 [20:06:04] - support switched from [nSignals x y] to [x y nSignals]. + % 2021.04.18 [14:42:53] - Updated to make imrect the default method of selecting the coordinates. % TODO % @@ -43,11 +44,20 @@ elseif isempty(options.inputCoords) %figure(102020); colormap gray; thisFrame = squeeze(inputMatrix(:,:,round(end/2))); - [coords] = getCropCoords(thisFrame) + % [coords] = getCropCoords(thisFrame) + + h = subfxn_getImRect(thisFrame,'Select a region'); + p = round(wait(h)); + + % Get the x and y corner coordinates as integers + coords(1) = p(1); %xmin + coords(2) = p(2); %ymin + coords(3) = p(1)+p(3); %xmax + coords(4) = p(2)+p(4); %ymax else coords = options.inputCoords; end - display('cropping matrix...'); + disp('cropping matrix...'); switch options.cropOrNaN case 'NaN' rowLen = size(inputMatrix,1); @@ -75,7 +85,8 @@ imagesc(thisFrame); axis image; colormap parula; - title('select region') + title('Select a region. Double click region to continue.') + box off; % Use ginput to select corner points of a rectangular % region by pointing and clicking the subject twice @@ -92,4 +103,19 @@ % Display the subsetted image with appropriate axis ratio figure(9);subplot(1,2,2);imagesc(thisFrameCropped); axis image; colormap gray; title('cropped region');drawnow; +end +function h = subfxn_getImRect(thisFrame,titleStr) + % close(142568) + figure(9); + clf + subplot(1,2,1); + imagesc(thisFrame); + axis image; + % colormap parula; + title('select region') + + h = imrect(gca); + addNewPositionCallback(h,@(p) title([titleStr 10 mat2str(p,3)])); + fcn = makeConstrainToRectFcn('imrect',get(gca,'XLim'),get(gca,'YLim')); + setPositionConstraintFcn(h,fcn); end \ No newline at end of file diff --git a/ciapkg/image/filterImages.m b/ciapkg/image/filterImages.m index a4a6c48..146cc33 100644 --- a/ciapkg/image/filterImages.m +++ b/ciapkg/image/filterImages.m @@ -161,21 +161,27 @@ display('done!'); if options.makePlots==1 - [figHandle figNo] = openFigure(99, ''); - % - subplot(2,1,1) - hist(imageSizes,round(logspace(0,log10(max(imageSizes))))); - box off;title('distribution of IC sizes');xlabel('area (px^2)');ylabel('count'); - set(gca,'xscale','log'); - h = findobj(gca,'Type','patch'); - set(h,'FaceColor',[0 0 0],'EdgeColor','w'); - % - subplot(2,1,2) - hist(find(valid==0),round(logspace(0,log10(max(find(valid==0)))))); - box off;title('rank of removed ICs');xlabel('rank');ylabel('count'); - set(gca,'xscale','log') - h = findobj(gca,'Type','patch'); - set(h,'FaceColor',[0 0 0],'EdgeColor','w'); + try + [figHandle figNo] = openFigure(99, ''); + % + subplot(2,1,1) + hist(imageSizes,round(logspace(0,log10(max(imageSizes))))); + box off;title('distribution of IC sizes');xlabel('area (px^2)');ylabel('count'); + set(gca,'xscale','log'); + h = findobj(gca,'Type','patch'); + set(h,'FaceColor',[0 0 0],'EdgeColor','w'); + % + subplot(2,1,2) + hist(find(valid==0),round(logspace(0,log10(max(find(valid==0)))))); + box off;title('rank of removed ICs');xlabel('rank');ylabel('count'); + set(gca,'xscale','log') + h = findobj(gca,'Type','patch'); + set(h,'FaceColor',[0 0 0],'EdgeColor','w'); + catch err + disp(repmat('@',1,7)) + disp(getReport(err,'extended','hyperlinks','on')); + disp(repmat('@',1,7)) + end end end % for i = 1:nImages diff --git a/ciapkg/io/loadMovieList.m b/ciapkg/io/loadMovieList.m index fed47c0..51dab4b 100644 --- a/ciapkg/io/loadMovieList.m +++ b/ciapkg/io/loadMovieList.m @@ -50,9 +50,12 @@ % 2020.08.31 [15:47:49] - Add option to suppress warnings. % 2020.10.19 [12:11:14] - Improved comments and options descriptions. % 2021.02.15 [11:55:36] - Fixed loading HDF5 datasetname that has only a single frame, loadMovieList would ask for 3rd dimension information that did not exist. + % 2021.06.21 [10:22:32] - Added support for Bio-Formats compatible files, specifically Olympus (OIR) and Zeiss (CZI, LSM). % TODO % OPEN + % Bio-Formats + % Allow outputting as a [x y c t] matrix to allow support for multiple color channels without needing to re-read file multiple times. % Determine file type by properties of file instead of extension (don't trust input...) % Remove all use of tmpMovie.... % Add ability to degrade gracefully with HDF5 dataset names, so try several backup datasetnames if one doesn't work. @@ -65,7 +68,12 @@ % ======================== % Cell array of str: list of supported file types, in general DO NOT change. - options.supportedTypes = {'.h5','.hdf5','.nwb','.tif','.tiff','.avi','.isxd'}; + options.supportedTypes = {'.h5','.hdf5','.tif','.tiff','.avi',... + '.nwb',... % Neurodata Without Borders format + '.isxd',... % Inscopix format + '.oir',... % Olympus formats + '.czi','.lsm'... % Zeiss formats + }; % Str: movie type. options.movieType = 'tiff'; % Str: hierarchy name in HDF5 file where movie data is located. @@ -106,8 +114,12 @@ options.useH5info = 1; % Int: [] = do nothing, 1-3 indicates R,G,B channels to take from multicolor RGB AVI options.rgbChannel = []; + % Int: Bio-Formats series number to load. + options.bfSeriesNo = 1; + % Int: Bio-Formats channel number to load. + options.bfChannelNo = 1; % get options - options = getOptions(options,varargin); + options = ciapkg.io.getOptions(options,varargin); % unpack options into current workspace % fn=fieldnames(options); % for i=1:length(fn) @@ -302,6 +314,22 @@ dims.two(iMovie) = xyDims(2); dims.three(iMovie) = nFrames; tmpFrame = inputMovieIsx.get_frame_data(0); + case 'bioformats' + bfreaderTmp = bfGetReader(thisMoviePath); + omeMeta = bfreaderTmp.getMetadataStore(); + stackSizeX = omeMeta.getPixelsSizeX(0).getValue(); % image width, pixels + stackSizeY = omeMeta.getPixelsSizeY(0).getValue(); % image height, pixels + stackSizeZ = omeMeta.getPixelsSizeZ(0).getValue(); + nFrames = stackSizeZ; + xyDims = [stackSizeY stackSizeX]; + dims.x(iMovie) = xyDims(1); + dims.y(iMovie) = xyDims(2); + dims.z(iMovie) = nFrames; + dims.one(iMovie) = xyDims(1); + dims.two(iMovie) = xyDims(2); + dims.three(iMovie) = nFrames; + tmpFrame = bfGetPlane(bfreaderTmp, 1); + nChannels = omeMeta.getChannelCount(0); end if isempty(options.loadSpecificImgClass) imgClass = class(tmpFrame); @@ -684,6 +712,67 @@ iframe = iframe + 1; end % ======================== + case 'bioformats' + % Setup movie class + bfreaderTmp = bfGetReader(thisMoviePath); + omeMeta = bfreaderTmp.getMetadataStore(); + stackSizeX = omeMeta.getPixelsSizeX(0).getValue(); % image width, pixels + stackSizeY = omeMeta.getPixelsSizeY(0).getValue(); % image height, pixels + stackSizeZ = omeMeta.getPixelsSizeZ(0).getValue(); + nChannels = omeMeta.getChannelCount(0); + + nFramesHere = stackSizeZ; + xyDims = [stackSizeX stackSizeY]; + + if isempty(thisFrameList) + nFrames = nFramesHere; + framesToGrab = 1:nFrames; + else + nFrames = length(thisFrameList); + framesToGrab = thisFrameList; + end + vidHeight = xyDims(1); + vidWidth = xyDims(2); + + % Preallocate movie structure. + tmpMovie = zeros(vidHeight, vidWidth, nFrames, imgClass); + + % Read one frame at a time. + reverseStr = ''; + iframe = 1; + nFrames = length(framesToGrab); + + % Read in movie data + tmpMovie = bfopen(thisMoviePath); + + % bfopen returns an n-by-4 cell array, where n is the number of series in the dataset. If s is the series index between 1 and n: + % The data{s, 1} element is an m-by-2 cell array, where m is the number of planes in the s-th series. If t is the plane index between 1 and m: + % The data{s, 1}{t, 1} element contains the pixel data for the t-th plane in the s-th series. + % The data{s, 1}{t, 2} element contains the label for the t-th plane in the s-th series. + % The data{s, 2} element contains original metadata key/value pairs that apply to the s-th series. + % The data{s, 3} element contains color lookup tables for each plane in the s-th series. + % The data{s, 4} element contains a standardized OME metadata structure, which is the same regardless of the input file format, and contains common metadata values such as physical pixel sizes - see OME metadata below for examples. + + % Frame information + frameInfo = tmpMovie{options.bfSeriesNo, 1}(:,2); + + % Grab just the movie frames and convert from cell to matrix. + tmpMovie = tmpMovie{options.bfSeriesNo, 1}(:,1); + tmpMovie = cat(3,tmpMovie{:}); + + % Only keep single channel if more than 1 channel in an image. + if nChannels>1 + try + chanKeepIdx = cell2mat(cellfun(@(x) str2num(cell2mat(regexp(x,'(?<=C\?=|C=)\d+(?=/)','match'))),frameInfo,'UniformOutput',false)); + chanKeepIdx = chanKeepIdx==options.bfChannelNo; + tmpMovie = tmpMovie(:,:,chanKeepIdx); + catch err + disp(repmat('@',1,7)) + disp(getReport(err,'extended','hyperlinks','on')); + disp(repmat('@',1,7)) + end + end + % ======================== otherwise % let's just not deal with this for now return; @@ -840,16 +929,20 @@ return; end % files are assumed to be named correctly (lying does no one any good) - if strcmp(ext,'.h5')||strcmp(ext,'.hdf5') + if any(strcmp(ext,{'.h5','.hdf5'})) movieType = 'hdf5'; elseif strcmp(ext,'.nwb') movieType = 'hdf5'; - elseif strcmp(ext,'.tif')||strcmp(ext,'.tiff') + elseif any(strcmp(ext,{'.tif','.tiff'})) movieType = 'tiff'; elseif strcmp(ext,'.avi') movieType = 'avi'; - elseif strcmp(ext,'.isxd') + elseif strcmp(ext,'.isxd') % Inscopix file format movieType = 'isxd'; + elseif strcmp(ext,'.oir') % Olympus file format + movieType = 'bioformats'; + elseif any(strcmp(ext,{'.czi','.lsm'})) % Zeiss file format + movieType = 'bioformats'; else movieType = ''; supported = 0; diff --git a/ciapkg/io/manageMiji.m b/ciapkg/io/manageMiji.m index 7e6a843..726d5d6 100644 --- a/ciapkg/io/manageMiji.m +++ b/ciapkg/io/manageMiji.m @@ -8,7 +8,7 @@ function manageMiji(varargin) % % changelog - % + % 2021.06.20 [00:20:38] - Add support for setting up ImageJ along with closing all windows to future proof any changes to those calls. % TODO % @@ -33,7 +33,7 @@ function manageMiji(varargin) % If MIJ class not loaded, load Miji.m. if exist('MIJ','class')~=8 % Load Miji so paths added to javaclasspath('-dynamic') - currP=pwd; + currP = pwd; % Miji; Miji(false); cd(currP); @@ -60,8 +60,11 @@ function manageMiji(varargin) end % If Miji.m not in path, ask user - if exist('Miji.m','file')~=2 - modelAddOutsideDependencies('miji'); + if exist('MIJ','class')~=8 + else + if exist('Miji.m','file')~=2 + modelAddOutsideDependencies('miji'); + end end % First attempt to open Miji @@ -85,6 +88,37 @@ function manageMiji(varargin) disp(repmat('@',1,7)) end end + case 'setupImageJ' + % Sets up ImageJ by pointing Java path to the jar files. + + imagejPath = [ciapkg.getDirExternalPrograms filesep 'imagej']; + pathsToAdd = {[imagejPath filesep 'mij.jar'],[imagejPath filesep 'ij.jar']}; + nPaths = length(pathsToAdd); + for i = 1:nPaths + thisPath = pathsToAdd{i}; + disp('Loading MIJI + ImageJ.') + fprintf('Adding to Java path: %s\n',thisPath); + javaaddpath(thisPath); + end + case 'closeAllWindows' + % Closes all open windows but leaves ImageJ running. + + % MIJ.run('Close All Without Saving'); + % MIJ.closeAllWindows; + allClosedFlag = 0 + nImagesToBreak = 20; + imageNo = 1 + while allClosedFlag==0 + try + MIJ.getListImages + MIJ.run('Close') + catch + allClosedFlag = 1; + end + imageNo = imageNo + 1; + end + MIJ.closeAllWindows; + % MIJ.exit; case 'exit' try MIJ.exit; diff --git a/ciapkg/io/modelAddOutsideDependencies.m b/ciapkg/io/modelAddOutsideDependencies.m index e67cea7..f3a5bc7 100644 --- a/ciapkg/io/modelAddOutsideDependencies.m +++ b/ciapkg/io/modelAddOutsideDependencies.m @@ -9,7 +9,8 @@ % changelog % 2019.10.15 [12:29:30] - Added flag to prevent recursive loop between resetMiji and modelAddOutsideDependencies. - % 2021.02.01 [15:19:40] - Update `_external_programs` to call ciapkg.getDirExternalPrograms() to standardize call across all functions. + % 2021.02.01 [??15:19:40] - Update `_external_programs` to call ciapkg.getDirExternalPrograms() to standardize call across all functions. + % 2021.06.20 [00:20:12] - Updated to add support for ImageJ call instead of Fiji. % TODO % @@ -31,6 +32,17 @@ success = 0; switch dependencyName case 'miji' + if exist('MIJ','class')==8 + disp('Miji loaded!') + return; + else + manageMiji('startStop','setupImageJ'); + if exist('MIJ','class')==8 + disp('Miji loaded!') + return; + end + end + if exist('Miji.m','file')==2 display(['Miji located in: ' which('Miji.m')]); % Miji is loaded, continue @@ -61,7 +73,7 @@ if exist('pathToMiji','var')==0 if exist(options.defaultExternalProgramDir,'dir')==7 loadPathHere = options.defaultExternalProgramDir; - loadStr = ['Enter path to Miji.m in Fiji (likely in "scripts" folder, e.g. calciumImagingAnalysis\' options.defaultExternalProgramDir '\Fiji.app\scripts)']; + loadStr = ['Enter path to Miji.m in Fiji (likely in "scripts" folder, e.g. ' filesep ciapkg.pkgName options.defaultExternalProgramDir '\Fiji.app\scripts)']; else loadPathHere = '\.'; loadStr = ['Enter path to Miji.m in Fiji (likely in "scripts" folder, e.g. \Fiji.app\scripts)']; diff --git a/ciapkg/io/saveMatrixToFile.m b/ciapkg/io/saveMatrixToFile.m index 07d1644..63e329a 100644 --- a/ciapkg/io/saveMatrixToFile.m +++ b/ciapkg/io/saveMatrixToFile.m @@ -9,7 +9,7 @@ % % changelog - % + % 2021.04.24 [16:00:01] - updated TIFF saving to add support for export of multi-channel color timeseries TIFF stack if in format [x y C t] where x,y = width/height, C = RGB channels, t = frames % TODO % Add checking of data size so tiff can be automatically switched @@ -82,6 +82,10 @@ case 'tiff' tiffOptions.comp = 'no'; tiffOptions.overwrite = true; + if length(size(inputMatrix))==4 + tiffOptions.color = true; + disp('Saving TIFF as color timeseries stack.'); + end fprintf('Saving to: %s\n',savePath); saveastiff(inputMatrix, savePath, tiffOptions); case 'hdf5' diff --git a/ciapkg/motion_correction/computeManualMotionCorrection.m b/ciapkg/motion_correction/computeManualMotionCorrection.m index 17dd694..76794b3 100644 --- a/ciapkg/motion_correction/computeManualMotionCorrection.m +++ b/ciapkg/motion_correction/computeManualMotionCorrection.m @@ -15,7 +15,9 @@ % changelog % 2020.04.07 [19:39:24] - Updated to allow just using the register aspect of the function. Also made registering callback based. % 2020.04.08 [10:35:49] - Added support for rotation. - % 2020.05.28 [08:48:57] - Slight update + % 2020.05.28 [08:48:57] - Slight update. + % 2021.04.16 [10:31:29] - Add default gamma option. + % 2021.04.27 [16:26:14] - Update to fix issue of prior figure (even after clf) maintaining previous key press and exiting, causing uiwait to fail. % TODO % Add ability to auto-crop if inputs are not of the right size them convert back to correct size after manual correction % inputRegisterImage - [x y nCells] - Image to register to. @@ -35,6 +37,10 @@ options.registerUseOutlines = 1; % Cell array of matrices: cell array of {[x y z]} matrices that should match each Z dimension in inputImages options.altInputImages = {}; + % Str: max or mean + options.cellCombineType = 'max'; + % Float: default gamma. + options.gammaCorrection = 1; % get options options = getOptions(options,varargin); % display(options) @@ -58,8 +64,15 @@ options.altInputImages = inputImages; else switchInputImagesBack = 0; - end - inputImages = cellfun(@(x) nanmax(x,[],3),inputImages,'UniformOutput',false); + end + switch options.cellCombineType + case 'max' + inputImages = cellfun(@(x) nanmax(x,[],3),inputImages,'UniformOutput',false); + case 'mean' + inputImages = cellfun(@(x) nanmean(x,3),inputImages,'UniformOutput',false); + otherwise + inputImages = cellfun(@(x) nanmax(x,[],3),inputImages,'UniformOutput',false); + end inputImages = cat(3,inputImages{:}); disp(['inputImages: ' num2str(size(inputImages))]) else @@ -141,12 +154,14 @@ inputRegisterImageOutlinesOriginal = inputRegisterImageOutlines; [figHandle figNo] = openFigure(options.translationFigNo, ''); + % Force current character to be a new figure. + set(gcf,'currentch','3'); clf % normalize input marker image inputImages = normalizeVector(single(inputImages),'normRange','zeroToOne'); inputImagesOriginal = inputImages; - gammaCorrection = 1; - gammaCorrectionRef = 1; + gammaCorrection = options.gammaCorrection; + gammaCorrectionRef = options.gammaCorrection; inputImages = imadjust(inputImages,[],[],gammaCorrection); inputRegisterImage = imadjust(inputRegisterImage,[],[],gammaCorrectionRef); continueRegistering = 1; @@ -157,7 +172,7 @@ set(figHandle, 'KeyPressFcn', @(source,eventdata) subfxnRespondUser(source,eventdata)); figure(figHandle) rgbImage = subfxncreateRgbImg(); - imgTitleFxn = @(imgNo,imgN,gammaCorrection,gammaCorrectionRef,translationVector,rotationVector) sprintf('Image %d/%d\nup/down/left/right arrows for translation | "A" = rotate left, "S" = rotate right | f to finish\n1/2 keys for image gamma down/up | gamma = %0.3f | gamma_r = %0.3f | translation %d %d | rotation %d\npurple = reference image, green = image to manually translate',imgNo,imgN,gammaCorrection,gammaCorrectionRef,translationVector,rotationVector); + imgTitleFxn = @(imgNo,imgN,gammaCorrection,gammaCorrectionRef,translationVector,rotationVector) sprintf('Image %d/%d\nup/down/left/right arrows for translation | "A" = rotate left, "S" = rotate right | f to finish\n1/2 keys for image gamma down/up | gamma = %0.3f | gamma(ref) = %0.3f | translation %d %d | rotation %d\npurple = reference image, green = image to manually translate',imgNo,imgN,gammaCorrection,gammaCorrectionRef,translationVector,rotationVector); imgTitle = imgTitleFxn(imgNo,imgN,gammaCorrection,gammaCorrectionRef,translationVector,rotationVector); imgHandle = imagesc(rgbImage); @@ -226,7 +241,8 @@ function subfxnDrawImg() if gammaCorrection<=1 gDelta = gDelta/10; else - gammaCorrection = round(gammaCorrection); + gDelta = gDelta/5; + % gammaCorrection = gammaCorrection+round(gammaCorrection-round(gammaCorrection)); end gammaCorrection = gammaCorrection+gDelta; if gammaCorrection<0 diff --git a/ciapkg/movie_processing/normalizeMovie.m b/ciapkg/movie_processing/normalizeMovie.m index bdc7808..db83c84 100644 --- a/ciapkg/movie_processing/normalizeMovie.m +++ b/ciapkg/movie_processing/normalizeMovie.m @@ -13,6 +13,7 @@ % 2019.10.08 [09:14:33] - Add option for user to input crop coordinates in case they have a NaN or other border from motion correction, so that does not affect spatial filtering. User can also now input a char for inputMovie and have it load within the function to help reduce memory overhead % 2019.10.29 [13:51:04] - Added support for parallel.pool.Constant when PCT auto-start parallel pool disabled. % 2021.01.15 [21:09:55] - Moved detrend support into normalizeMovie. + % 2021.06.02 [20:04:49] - Detrend movie now uses nanmean to get around issues in motion corrected videos. % TODO % @@ -717,12 +718,12 @@ function subfxnDetrend() %Get dimension information about 3D movie matrix [inputMovieX, inputMovieY, inputMovieZ] = size(inputMovie); - frameMeanInputMovie = squeeze(mean(inputMovie,[1 2])); + frameMeanInputMovie = squeeze(nanmean(inputMovie,[1 2])); trendVals = frameMeanInputMovie - detrend(frameMeanInputMovie,option.detrendDegree); % meanInputMovie = detrend(frameMeanInputMovie,0); - meanInputMovie = mean(frameMeanInputMovie); + meanInputMovie = nanmean(frameMeanInputMovie); nFramesToNormalize = options.maxFrame; nFrames = nFramesToNormalize; diff --git a/ciapkg/video/createMovieFromVector.m b/ciapkg/video/createMovieFromVector.m index 2ea0aad..816c528 100644 --- a/ciapkg/video/createMovieFromVector.m +++ b/ciapkg/video/createMovieFromVector.m @@ -9,7 +9,7 @@ % vectorMovie - 3D movie ([x y t]) with dimensions of movieDim and class of inputVector % changelog - % + % 2021.05.04 [09:29:19] - Users can now manually change value assigned to center line or signal. % TODO % @@ -20,6 +20,10 @@ options.windowSize = [-30:30]; % movie height downsample size options.secondDimDownsample = 5; + % Float: numbers between 0 to 1, value to make for signal + options.signalValue = 0.5; + % Float: numbers between 0 to 1, value to make for center line + options.centerLineValue = 1; % get options options = getOptions(options,varargin); % display(options) @@ -41,7 +45,7 @@ nStims = length(windowSize); if ~isempty(options.normalizeVector) - display('Normalizing vector...') + disp('Normalizing vector...') inputVector = normalizeVector(inputVector,'normRange',options.normalizeVector); end @@ -66,12 +70,12 @@ relativeStimValue = round(inputVector(frameVectorIdx(thisFrameVectorNo))*movieDimY); end % add relative (to max) value of vector to movie - vectorMovie(1:relativeStimValue,thisFrameVectorNo,frameNo) = 0.05; + vectorMovie(1:relativeStimValue,thisFrameVectorNo,frameNo) = options.signalValue; end % resize vector movie to match movie dimensions given vectorMovie(:,:,frameNo) = imresize(vectorMovie(:,1:length(frameVectorIdx),frameNo),[movieDimY movieDim(2)],'bilinear'); - vectorMovie(:,round(end/2),frameNo) = 1; + vectorMovie(:,round(end/2),frameNo) = options.centerLineValue; reverseStr = cmdWaitbar(frameNo,nFrames,reverseStr,'inputStr','creating matrix: ','waitbarOn',1,'displayEvery',5); end diff --git a/ciapkg/view/playMovie.m b/ciapkg/view/playMovie.m index 852acb8..5934c7b 100644 --- a/ciapkg/view/playMovie.m +++ b/ciapkg/view/playMovie.m @@ -4,7 +4,9 @@ % started 2013.11.09 [10:39:50] % % inputs - % inputMovie - [X Y Z] matrix of X,Y height/width and Z frames + % inputMovie - either grayscale or color movie matrix: + % grayscale: [x y t] matrix of x,y height/width and t frames + % RGB: [x y C t] matrix of x,y height/width, t frames, and C color channel (3 as RGB) % options % fps - % extraMovie - extra movie to play, [X Y Z] matrix of X,Y height/width and Z frames @@ -29,6 +31,7 @@ % 2021.01.14 [20:12:10] - Update passing of HDF5 dataset name to ciapkg.io.readFrame. % 2021.01.17 [19:21:12] - Integrated contrast sliders directly into the main GUI so users don't have to open up a separate GUI. Make GUI sliders thinner. % 2021.02.05 [16:25:12] - Added feature to sub-sample movie to make display run faster for larger movies. + % 2021.04.23 [13:05:29] - Add support for playing RGB movie of dimension [x y C t] if input directly as matrix. % ======================== % options @@ -150,7 +153,11 @@ else inputMovieIsChar = 0; inputMovieDims = size(inputMovie); - nFramesOriginal = inputMovieDims(3); + if length(inputMovieDims)==4 + nFramesOriginal = inputMovieDims(4); + else + nFramesOriginal = inputMovieDims(3); + end readMovieChunks = 0; end @@ -167,7 +174,11 @@ % ========================================== if options.nFrames==0 - nFrames = size(inputMovie,3); + if length(inputMovieDims)==4 + nFrames = size(inputMovie,4); + else + nFrames = size(inputMovie,3); + end else nFrames = options.nFrames; end @@ -261,7 +272,11 @@ [tmpFrame] = ciapkg.io.readFrame(inputMovie,1,'movieFileID',movieFileID,'inputMovieDims',inputMovieDims,'inputDatasetName',options.inputDatasetName); % tmpFrame = loadMovieList(inputMovie,'inputDatasetName',options.inputDatasetName,'displayInfo',0,'displayDiagnosticInfo',0,'displayWarnings',0,'frameList',1); else - tmpFrame = squeeze(inputMovie(:,:,1)); + if length(size(inputMovie))==4 + tmpFrame = squeeze(inputMovie(:,:,:,1)); + else + tmpFrame = squeeze(inputMovie(:,:,1)); + end end if sparseInputMovie==1 tmpFrame = full(tmpFrame); @@ -457,7 +472,11 @@ % thisFrame = subfxn_readMovieDisk(inputMovie,frame,movieType); [thisFrame] = ciapkg.io.readFrame(inputMovie,frame,'movieFileID',movieFileID,'inputMovieDims',inputMovieDims,'inputDatasetName',options.inputDatasetName); else - thisFrame = squeeze(inputMovie(:,:,frame)); + if length(size(inputMovie))==4 + thisFrame = squeeze(inputMovie(:,:,:,frame)); + else + thisFrame = squeeze(inputMovie(:,:,frame)); + end end if sparseInputMovie==1 thisFrame = full(thisFrame); @@ -523,7 +542,11 @@ ssm = options.subSampleMovie; set(montageHandle,'Cdata',thisFrame(1:ssm:end,1:ssm:end),'AlphaData',imAlpha(1:ssm:end,1:ssm:end)); else - set(montageHandle,'Cdata',thisFrame,'AlphaData',imAlpha); + if length(size(thisFrame))==3 + % set(montageHandle,'Cdata',squeeze(thisFrame(:,:,1)),'AlphaData',imAlpha); + else + set(montageHandle,'Cdata',thisFrame,'AlphaData',imAlpha); + end end if strcmp(options.colormapColor,'gray') set(axHandle,'color',[1 0 0]); diff --git a/ciapkg/view/viewVennDiagram.m b/ciapkg/view/viewVennDiagram.m index 0c79cb4..265aa06 100644 --- a/ciapkg/view/viewVennDiagram.m +++ b/ciapkg/view/viewVennDiagram.m @@ -124,6 +124,7 @@ function viewVennDiagram(circleAreas,overlapAreas,totalArea,varargin) % Plot text indicating the exact numbers for each area of overlap if options.displayText==1 + rdDgts = options.roundDigits; % str1 = sprintf('%d | %0.1f | %0.1f',idPairsFixed(idPairNo,1), circleAreasOriginal(1),circleAreas(1)); % str2 = sprintf('%d | %0.1f | %0.1f',idPairsFixed(idPairNo,2), circleAreasOriginal(2),circleAreas(2)); % str3 = sprintf('%d | %0.1f | %0.1f',idPairsFixed(idPairNo,3), circleAreasOriginal(3),circleAreas(3)); diff --git a/docs/docs/acknowledgments.md b/docs/docs/acknowledgments.md index e6dc51e..8647c23 100644 --- a/docs/docs/acknowledgments.md +++ b/docs/docs/acknowledgments.md @@ -1,4 +1,4 @@ -## Acknowledgments +# Acknowledgments Thanks to Jones G. Parker, PhD (
—Setup—
+
+{!install.md!}
+{!install_alt.md!}
+{!example_data.md!}
+{!dependencies.md!}
+
+## —Repository—
+{!data.md!}
+{!notes.md!}
+{!organization.md!}
+
+## —Processing data—
+{!pipeline_overview.md!}
+{!pipeline_detailed.md!}
+{!pipeline_animal_tracking.md!}
+
+## —API—
+{!api_example_pipeline.md!}
+{!api_ciapkg.md!}
+
+## —Help—
+{!help_issues.md!}
+{!help_inscopix.md!}
+{!help_nwb.md!}
+{!help_contrast.md!}
+{!help_analysis_methods.md!}
+{!help_large_movie_analysis.md!}
+{!help_stripe_removal.md!}
+{!help_snr.md!}
+{!help_spatial_filtering.md!}
+{!help_temporal_downsampling.md!}
+{!help_motion_correction.md!}
+{!help_cell_extraction.md!}
+{!help_manual_cell_sorting.md!}
+{!help_cross_session_alignment.md!}
+{!help_animal_tracking.md!}
+
+## —Misc—
+{!acknowledgments.md!}
+{!references.md!}
+{!questions.md!}
+{!license.md!}
\ No newline at end of file
diff --git a/docs/docs/api_ciapkg.md b/docs/docs/api_ciapkg.md
new file mode 100644
index 0000000..907a847
--- /dev/null
+++ b/docs/docs/api_ciapkg.md
@@ -0,0 +1,53 @@
+# `{{ site.name }}` functions within `{{ code.package }}` sub-package.
+
+`{{ site.name }}` contains many functions for imaging analysis, from processing videos to wrappers for running cell extraction algorithms and GUIs for visualizing movies and evaluating cell extraction outputs. Several are detailed below. For each users can visualize options with `help FUN` or `edit FUN`. If attempting to load a non-package function (e.g. it does not start with `{{ code.package }}`), then append `{{ code.package }}.api.`, e.g. `playMovie` would become `{{ code.package }}.api.playMovie`. Alternatively, load all the functions into the workspace with `import ciapkg.api.*`.
+
+## Visualizing movies
+
+`playMovie`
+
+`ciapkg.io.loadMovie` or `loadMovieList`
+
+`createImageOutlineOnMovie`
+
+`createSignalBasedMovie`
+
+`ciapkg.io.readFrame`
+
+## Get movie information
+
+`ciapkg.io.getMovieInfo`
+
+## Sorting cells
+
+`signalSorter`
+
+## Pre-processing
+
+`ciapkg.demo.runPreprocessing`
+
+`downsampleHdf5Movie`
+
+`removeStripsFromMovie`
+
+`turboregMovie`
+
+`dfofMovie`
+
+`downsampleMovie`
+
+`normalizeMovie`
+
+## Cell extraction
+
+- _PCA-ICA_ - `ciapkg.signal_extraction.runPcaIca`.
+- _CNMF_ - ``.
+- _CNMF-E_ - ``.
+- _EXTRACT_ - ``.
+- _ROI_ - ``.
+
+## Cross-session alignment
+
+`matchObjBtwnTrials`
+
+`createMatchObjBtwnTrialsMaps`
\ No newline at end of file
diff --git a/docs/docs/api_example_pipeline.md b/docs/docs/api_example_pipeline.md
new file mode 100644
index 0000000..146d151
--- /dev/null
+++ b/docs/docs/api_example_pipeline.md
@@ -0,0 +1,196 @@
+# Example `{{ site.name }}` pipeline via the command line.
+
+Below is an example `{{ site.name }}` pipeline using the command line for those that do not want to use the class or want to create their own custom batch analyses. It assumes you have already run `example_downloadTestData` to download the example test data and have MATLAB path set to the `{{ site.name }}` root directory.
+
+Can also access the pipeline by typing `edit ciapkg.demo.cmdLinePipeline` into the MATLAB command window or run by typing in `ciapkg.demo.cmdLinePipeline;`.
+
+```MATLAB
+% Running {{ site.name }} from MATLAB command line/window
+
+%% Load {{ site.name }} functions
+loadBatchFxns();
+
+%% Load movie to analyze
+inputMovie = loadMovieList([ciapkg.getDir() filesep 'data' filesep '2014_04_01_p203_m19_check01' filesep 'concat_recording_20140401_180333.h5']);
+```
+
+```MATLAB
+%% Visualize slice of the movie
+playMovie(inputMovie(:,:,1:500));
+% Alternatively, visualize by entering the file path
+playMovie(inputMoviePath);
+```
+
+```MATLAB
+%% Downsample input movie if need to
+inputMovieD = downsampleMovie(inputMovie,'downsampleDimension','space','downsampleFactor',4);
+playMovie(inputMovie,'extraMovie',inputMovieD);
+
+% Alternatively, if you have Inscopix ISXD files, downsample by reading segments from disk using.
+moviePath = 'PATH_TO_ISXD';
+opts.maxChunkSize = 5000; % Max chunk size in Mb to load into RAM.
+opts.downsampleFactor = 4; % How much to downsample original movie, set to 1 for no downsampling.
+convertInscopixIsxdToHdf5(moviePath,'options',opts);
+```
+
+```MATLAB
+%% Remove stripes from movie if needed
+% Show full filter sequence for one frame
+sopts.stripOrientation = 'both';
+sopts.meanFilterSize = 1;
+sopts.freqLowExclude = 10;
+sopts.bandpassType = 'highpass';
+removeStripsFromMovie(inputMovie(:,:,1),'options',sopts,'showImages',1);
+% Run on the entire movie
+removeStripsFromMovie(inputMovie,'options',sopts);
+```
+
+```MATLAB
+%% Get coordinates to crop
+[cropCoords] = getCropCoords(squeeze(inputMovie(:,:,1)));
+toptions.cropCoords = cropCoords;
+```
+
+```MATLAB
+%% Motion correction
+% Or have turboreg run manual correction
+toptions.cropCoords = 'manual';
+toptions.turboregRotation = 0;
+toptions.removeEdges = 1;
+toptions.pxToCrop = 10;
+% Pre-motion correction
+ toptions.complementMatrix = 1;
+ toptions.meanSubtract = 1;
+ toptions.meanSubtractNormalize = 1;
+ toptions.normalizeType = 'matlabDisk';
+% Spatial filter
+ toptions.normalizeBeforeRegister = 'divideByLowpass';
+ toptions.freqLow = 0;
+ toptions.freqHigh = 7;
+[inputMovie2, ~] = turboregMovie(inputMovie,'options',toptions);
+```
+
+```MATLAB
+%% Compare raw and motion corrected movies
+playMovie(inputMovie,'extraMovie',inputMovie2);
+```
+
+```MATLAB
+%% Run dF/F
+inputMovie3 = dfofMovie(single(inputMovie2),'dfofType','dfof');
+```
+
+```MATLAB
+%% Run temporal downsampling
+inputMovie3 = downsampleMovie(inputMovie3,'downsampleDimension','time','downsampleFactor',4);
+```
+
+```MATLAB
+%% Final check of movie before cell extraction
+playMovie(inputMovie3);
+```
+
+```MATLAB
+%% Run PCA-ICA cell extraction. CNMF-e, CNMF, ROI, and other cell-extraction algorithms are also available.
+nPCs = 300; nICs = 225;
+[PcaOutputSpatial, PcaOutputTemporal, PcaOutputSingularValues, PcaInfo] = run_pca(inputMovie3, nPCs, 'movie_dataset_name','/1');
+[IcaFilters, IcaTraces, IcaInfo] = run_ica(PcaOutputSpatial, PcaOutputTemporal, PcaOutputSingularValues, size(inputMovie3,1), size(inputMovie3,2), nICs, 'output_units','fl','mu',0.1,'term_tol',5e-6,'max_iter',1e3);
+IcaTraces = permute(IcaTraces,[2 1]);
+```
+
+```MATLAB
+%% Save outputs to NWB format
+saveNeurodataWithoutBorders(IcaFilters,{IcaTraces},'pcaica','pcaica.nwb');
+```
+
+Run CNMF or CNMF-e cell extraction.
+```MATLAB
+%% Run CNMF or CNMF-e cell extraction. Check each function with "edit" for options.
+numExpectedComponents = 225;
+cellWidth = 10;
+cnmfOptions.otherCNMF.tau = cellWidth/2; % expected width of cells
+
+% Run CNMF
+[cnmfAnalysisOutput] = computeCnmfSignalExtractionClass(movieList,numExpectedComponents,'options',cnmfOptions);
+
+% Run CNMF-e
+[cnmfeAnalysisOutput] = computeCnmfeSignalExtraction_batch(movieList{1},'options',cnmfeOptions);
+
+%% Save outputs to NWB format
+saveNeurodataWithoutBorders(cnmfAnalysisOutput.extractedImages,{cnmfAnalysisOutput.extractedSignals,cnmfAnalysisOutput.extractedSignalsEst},'cnmf','cnmf.nwb');
+saveNeurodataWithoutBorders(cnmfeAnalysisOutput.extractedImages,{cnmfeAnalysisOutput.extractedSignals,cnmfeAnalysisOutput.extractedSignalsEst},'cnmfe','cnmfe.nwb');
+```
+
+Run EXTRACT cell extraction.
+```MATLAB
+%% Run EXTRACT cell extraction. Check each function with "edit" for options.
+% Load default configuration
+extractConfig = get_defaults([]);
+
+outStruct = extractor(inputMovie,extractConfig);
+extractAnalysisOutput.filters = outStruct.spatial_weights;
+% permute so it is [nCells frames]
+extractAnalysisOutput.traces = permute(outStruct.temporal_weights, [2 1]);
+
+% Other run information if saving as a MAT-file.
+extractAnalysisOutput.info = outStruct.info;
+extractAnalysisOutput.config = outStruct.config;
+extractAnalysisOutput.info = outStruct.info;
+extractAnalysisOutput.userInputConfig = extractConfig;
+extractAnalysisOutput.opts = outStruct.config;
+
+%% Save outputs to NWB format
+saveNeurodataWithoutBorders(extractAnalysisOutput.filters,{extractAnalysisOutput.traces},'cnmf','cnmf.nwb');
+```
+
+```MATLAB
+%% Run signal sorting using matrix inputs
+[outImages, outSignals, choices] = signalSorter(IcaFilters,IcaTraces,'inputMovie',inputMovie3);
+```
+
+```MATLAB
+%% Run signal sorting using NWB
+[outImages, outSignals, choices] = signalSorter('pcaica.nwb',[],'inputMovie',inputMovie3);
+```
+
+```MATLAB
+%% Plot results of sorting
+figure;
+subplot(1,2,1);imagesc(max(IcaFilters,[],3));axis equal tight; title('Raw filters')
+subplot(1,2,2);imagesc(max(outImages,[],3));axis equal tight; title('Sorted filters')
+```
+
+```MATLAB
+%% Create an overlay of extraction outputs on the movie and signal-based movie
+[inputMovieO] = createImageOutlineOnMovie(inputMovie3,IcaFilters,'dilateOutlinesFactor',0);
+[signalMovie] = createSignalBasedMovie(IcaTraces,IcaFilters,'signalType','peak');
+```
+
+```MATLAB
+%% Play all three movies
+% Normalize all the movies
+movieM = cellfun(@(x) normalizeVector(x,'normRange','zeroToOne'),{inputMovie3,inputMovieO,signalMovie},'UniformOutput',false);
+playMovie(cat(2,movieM{:}));
+```
+
+```MATLAB
+%% Run cross-session alignment of cells
+% Create input images, cell array of [x y nCells] matrices
+inputImages = {day1Images,day2Images,day3Images};
+
+% options to change
+opts.maxDistance = 5; % distance in pixels between centroids for them to be grouped
+opts.trialToAlign = 1; % which session to start alignment on
+opts.nCorrections = 1; %number of rounds to register session cell maps.
+opts.RegisTypeFinal = 2 % 3 = rotation/translation and iso scaling; 2 = rotation/translation, no iso scaling
+
+% Run alignment code
+[alignmentStruct] = matchObjBtwnTrials(inputImages,'options',opts);
+
+% Global IDs is a matrix of [globalID sessionID]
+% Each (globalID, sessionID) pair gives the within session ID for that particular global ID
+globalIDs = alignmentStruct.globalIDs;
+
+% View the cross-session matched cells, saved to `private\_tmpFiles` sub-folder.
+[success] = createMatchObjBtwnTrialsMaps(inputImages,alignmentStruct);
+```
\ No newline at end of file
diff --git a/docs/docs/blank.md b/docs/docs/blank.md
new file mode 100644
index 0000000..2ae31d9
--- /dev/null
+++ b/docs/docs/blank.md
@@ -0,0 +1,91 @@
+# {{ site.name }}
+
+
+
+[](https://github.com/bahanonu/calciumImagingAnalysis/releases/latest?style=flat-square&logo=appveyor)
+
+[](https://github.com/bahanonu/calciumImagingAnalysis/graphs/commit-activity?style=flat-square&logo=appveyor)
+
+ +
+
+
++ +Created by __Biafra Ahanonu, PhD__. + +`{{ site.name }}` (pronounced cheetah, formerly `calciumImagingAnalysis` [`CIAPKG`]) is a software package for analysis of one- and two-photon calcium imaging datasets. + +__Download the software at [https://github.com/bahanonu/calciumImagingAnalysis](https://github.com/bahanonu/calciumImagingAnalysis).__ + +
 +
+- This user guide contains instructions to setup, run, and troubleshoot `{{ site.name }}`.
+- Note: `{{ site.name }}` is a class (`{{ site.namelow }}` within MATLAB) with various GUIs to allow processing of calcium imaging data. In addition, users can access the underlying `{{ site.name }}` functions to make custom workflows. See [Custom command-line pipelines](api_example_pipeline.md).
+- Read my overview of calcium imaging analysis methods at [Calcium imaging cell identification and fluorescence activity trace reconstruction, part 1](https://bahanonu.com/brain/#c20181209).
+
+
+
+
+
+- This user guide contains instructions to setup, run, and troubleshoot `{{ site.name }}`.
+- Note: `{{ site.name }}` is a class (`{{ site.namelow }}` within MATLAB) with various GUIs to allow processing of calcium imaging data. In addition, users can access the underlying `{{ site.name }}` functions to make custom workflows. See [Custom command-line pipelines](api_example_pipeline.md).
+- Read my overview of calcium imaging analysis methods at [Calcium imaging cell identification and fluorescence activity trace reconstruction, part 1](https://bahanonu.com/brain/#c20181209).
+
+
+
++Movie processing, cell extraction, and analysis validation. +
+
+
+  +
+
+
+
+ {{ site.name }} cell sorting GUI +
+
+
+  +
+
+
+
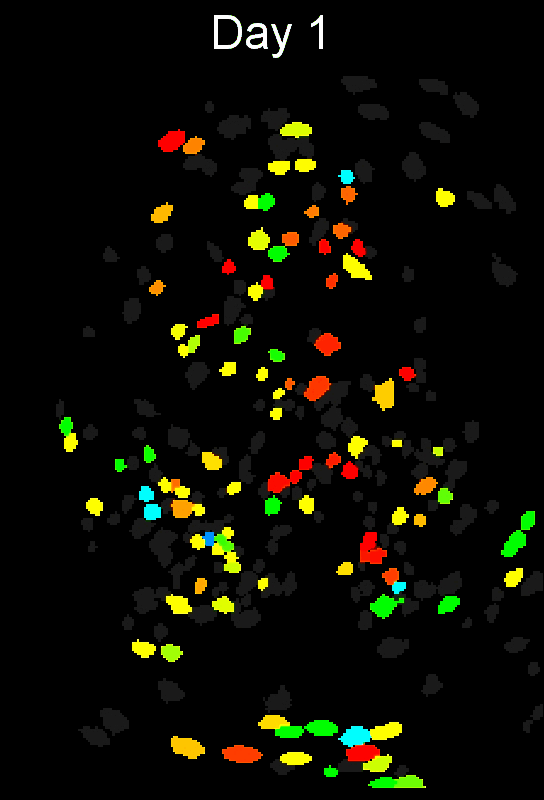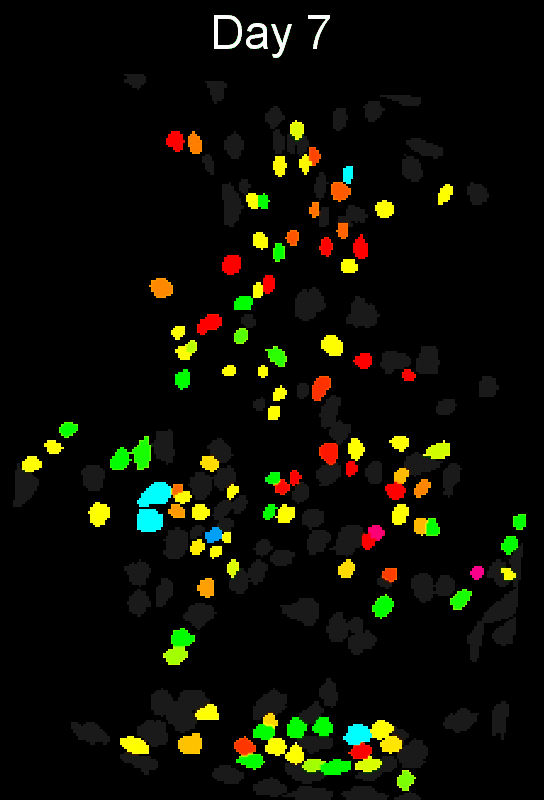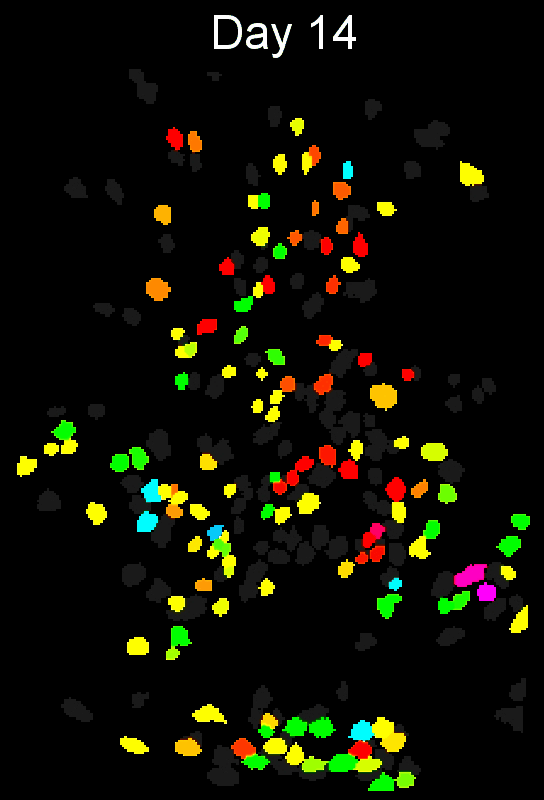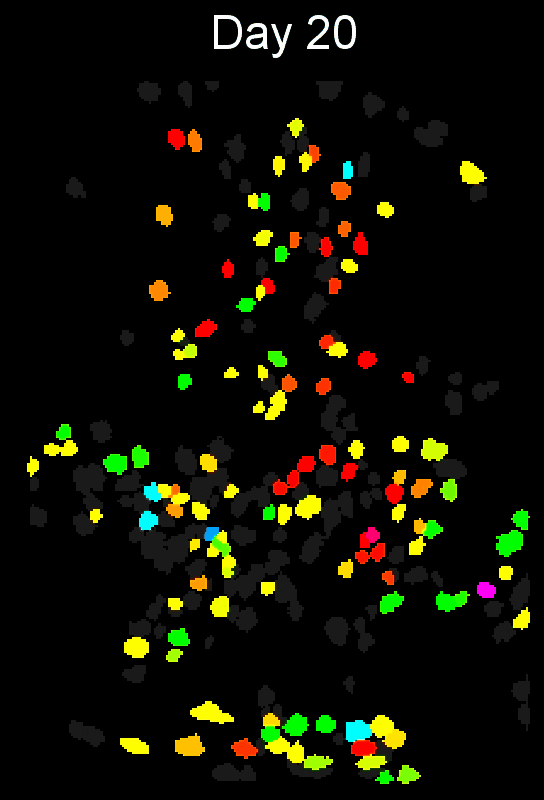
+ Stable cell alignment across imaging sessions. +
+
+
+  +
+
+
+
+
 +
+-  (starting 2020.09.16)
+-  (starting 2020.09.22)
+
\ No newline at end of file
diff --git a/docs/docs/example_data.md b/docs/docs/example_data.md
new file mode 100644
index 0000000..710ad4d
--- /dev/null
+++ b/docs/docs/example_data.md
@@ -0,0 +1,7 @@
+### Example data
+
+The `{{ site.name }}` repository contains several example one-photon calcium imaging movies from miniature microscope experiments in freely moving rodents.
+
+To download example data, run `loadDependencies` module (e.g. `obj.loadDependencies`) and select `Download test one-photon data.` option to download one-photon miniature microscope example datasets to use for testing `{{ site.name }}` preprocessing, cell extraction, and cell classification code. The data will be located in the `data` folder within the repository root directory.
+
+Else run `example_downloadTestData.m` from the MATLAB command line with the working directory set to the `{{ site.name }}` repository.
\ No newline at end of file
diff --git a/docs/docs/help_animal_tracking.md b/docs/docs/help_animal_tracking.md
index b31c80b..e0fc098 100644
--- a/docs/docs/help_animal_tracking.md
+++ b/docs/docs/help_animal_tracking.md
@@ -1,28 +1,51 @@
-# Tracking
+# Animal tracking
-Code for ImageJ and Matlab based image tracking.
+Code for ImageJ- and Matlab-based image tracking.
# ImageJ based tracking
-Functions needed (have entire `miniscope_analysis` loaded anyways):
+Functions needed (have entire `{{ site.name }}` pipeline loaded anyways to ensure all dependencies are met):
- `mm_tracking.ijm` is the tracking function for use in ImageJ, place in
-`plugins` folder.
-- `removeIncorrectObjs.m` is a function to clean-up the ImageJ output.
-- `createTrackingOverlayVideo` is a way to check the output from the
-tracking by overlaying mouse tracker onto the video.
+`plugins` folder. It is found in the `ciapkg\tracking` folder of the repository.
+- `removeIncorrectObjs()` is a function to clean-up the ImageJ output.
+- `createTrackingOverlayVideo()` is a function that allows users to check the output from the tracking by overlaying the mouse tracker onto the video.
## Instructions for ImageJ and Matlab
+Run `mm_tracking` from the `Plugins` menu of ImageJ. Several settings to check:
+
+- `number of session folders to analyze` - this will indicate how many movies you want to analyze. Depending on `analyze movies from a list file` setting, a GUI will appear asking you to select movies to analyze (preferably AVIs) or a text file with a list of movies.
+- `pixel to cm conversion, distance (cm)` - Make sure pixel to cm conversion indicates a measurable distance in the video, e.g. from one side of the box to another. The program will ask you to draw this distance to estimate pixels/cm.
+- `crop stack?` - keep this selected, as allow removal of parts of movie where the animal will not go, improving results.
+- `erode and dilate` - likely keep this selected, as it essentially makes sure the mouse is a solid object and smooths the tracking.
+- `analyze movies from a list file` - select this option if you have a text file with the location of each movie to be analyzed on a new line. Use this to analyze many movies in batch.
+- `hide loaded movie` - uncheck this to be able to visualize as ImageJ completes each step in the processing. Leave checked to improve speed of analysis.
+
Example screen after running `mm_tracking` within ImageJ, click to expand.
-
-Once ImageJ is finished, within Matlab run the following code (cleans up the ImageJ tracking by removing small objects and adding NaNs for missing frames along with making a movie to check output). Modify to point toward paths specific for your data.
+
+
+
+Once ImageJ is finished, within `MATLAB` run the following code (cleans up the ImageJ tracking by removing small objects and adding NaNs for missing frames along with making a movie to check output). Modify to point toward paths specific for your data.
```Matlab
% CSV file from imageJ and AVI movie path used in ImageJ
moviePath = 'PATH_TO_AVI_USED_IN_IMAEJ';
csvPath = 'PATH_TO_CSV_OUTPUT_BY_IMAGEJ';
% clean up tracking
[trackingTableFilteredCell] = removeIncorrectObjs(csvPath,'inputMovie',{moviePath});
+```
+
+## Example output from animal in open field during miniature microscope imaging
+
+
+Tracking of an animal over time (green = early in session, red = late in session).
+
+
+## Tracking video
+
+The tracking video can be used to quickly validate that the animal is being correctly tracked.
+
+```Matlab
% make tracking video
% frames to use as example check
nFrames=1500:2500;
@@ -31,12 +54,10 @@ inputMovie = loadMovieList(moviePath,'frameList',nFrames);
playMovie(inputTrackingVideo);
```
-## Example output from 2017_09_11_p540_m381_openfield01_091112017
-
-
+Overlay of tracking (red circle) on the mouse in a specific frame.

-# Matlab based tracking
+
diff --git a/docs/docs/help_cell_extraction.md b/docs/docs/help_cell_extraction.md
new file mode 100644
index 0000000..e91bffb
--- /dev/null
+++ b/docs/docs/help_cell_extraction.md
@@ -0,0 +1,2 @@
+# Cell extraction
+
diff --git a/docs/docs/help_contrast.md b/docs/docs/help_contrast.md
new file mode 100644
index 0000000..724fbe9
--- /dev/null
+++ b/docs/docs/help_contrast.md
@@ -0,0 +1,32 @@
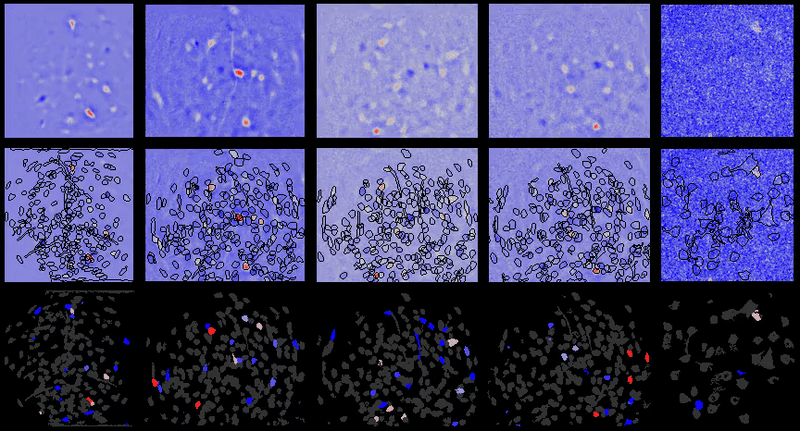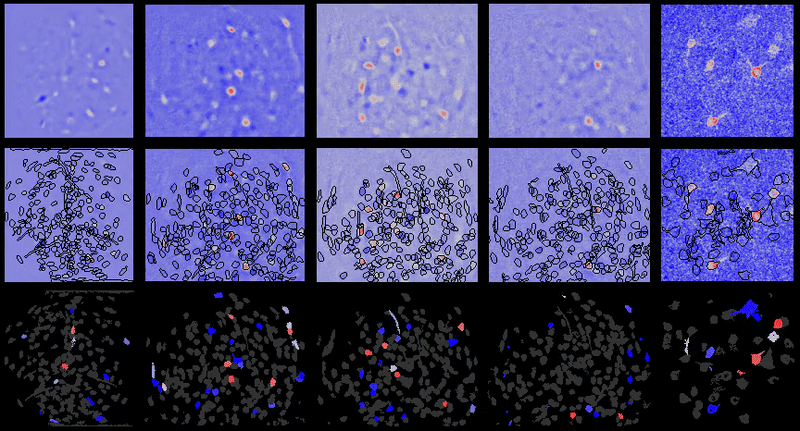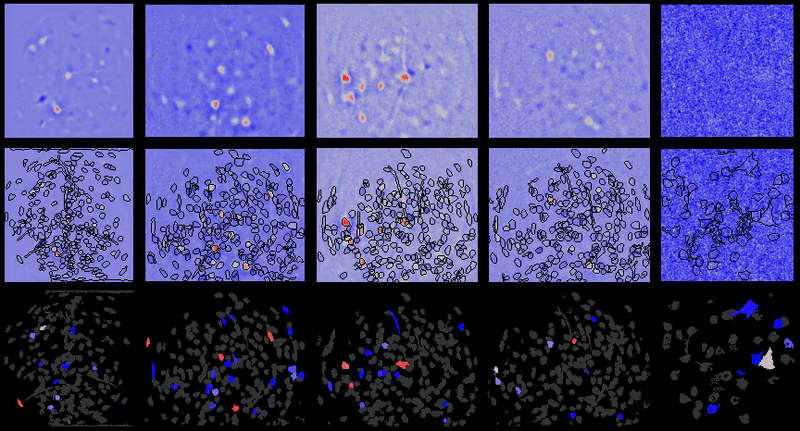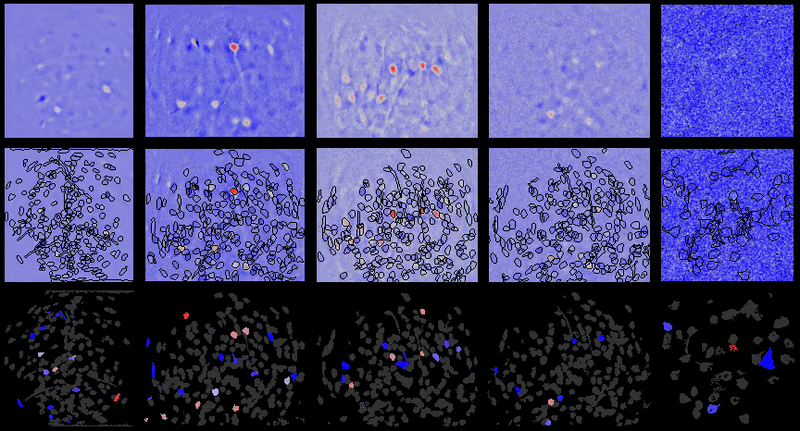
+# Interpreting displayed movies
+
+It may appear at times that `{{ site.name }}` has introduced noise into movies after processing. However, normally that noise is already there in the movie. In the case of one-photon miniature microscope movies that noise is often small relative to the baseline background fluorescence, hence not noticeable. However, after dF/F0 or spatial filtering users will be able to see the noise more easily as it will have a relatively greater magnitude compared to the signal now present in the movie.
+
+Further, in many cases the contrast is automatically adjusted in `{{ site.name }}` GUIs to boost likelihood users will see cells and other biologically relevant features (even though underlying matrix values are not changed), which can sometimes lead to the perception that noise is being added depending on viewing raw vs. dF/F0 or other preprocessed movies.
+
+## Example
+
+For example of what this looks like, take one of the example movies in the {{ site.name }} repository: `2014_04_01_p203_m19_check01`.
+
+### Raw movie
+
+A frame from the raw movie looks like:
+
+
+
+Applying a simple bandpass or highpass filter (to remove the low frequency background) leads to the below (keeping contrast/brightness the same as the raw movie image):
+
+
+
+However, if we adjust the contrast, we can now see some of the noise present in the higher frequency components of the raw movie that was otherwise obscured or would be hard to see in the raw movie with the high baseline present:
+
+
+
+### dF/F0
+Now compare to dF/F0 of that same raw movie without any motion correction, etc. we see the below:
+
+
+
+However, if you adjust the contrast, you get the below image, where the noise is much more pronounced:
+
+
\ No newline at end of file
diff --git a/docs/docs/help_large_movie_analysis.md b/docs/docs/help_large_movie_analysis.md
new file mode 100644
index 0000000..f619438
--- /dev/null
+++ b/docs/docs/help_large_movie_analysis.md
@@ -0,0 +1,44 @@
+# Analyzing large movies
+
+Some movies are larger than the available RAM on users analysis computer. Below are several ways that the underlying functions in `CIAPKG` can be used to analyze large movies.
+
+## Playing large movies from disk
+
+To directly and quickly visualize large or long movies from disk, directly input the movie path into `playMovie` as below.
+
+```Matlab
+% Full path to the movie
+inputMoviePath = 'path/to/movie.h5';
+
+playMovie(inputMoviePath);
+```
+
+Which will produce a GUI as below that will play the movie back.
+
+
+
+## ROI signal extraction
+The below code is an example ROI signal extraction for large movie in chunks from disk after analyzing a small chunk with PCA-ICA to obtain reference masks. Modify `inputMoviePath` to a full path to your movie (HDF5, TIF, AVI, and ISXD supported).
+
+```MATLAB
+% Full path to the movie
+inputMoviePath = 'path/to/movie.h5';
+
+%% =======PCA-ICA
+% Run PCA-ICA on only a subset of frames.
+% OPTIONS
+ % Vector of frames to analyze for PCA-ICA
+ framesToAnalyzePcaIca = 1:300;
+ % Number of PCs and ICs to request
+ nPCs = 250; nICs = 200;
+[pcaicaAnalysisOutput] = ciapkg.signal_extraction.runPcaIca(inputMoviePath,nPCs,nICs,'frameList',framesToAnalyzePcaIca,'mu',0.1,'max_iter',1e3);
+
+%% =======ROI extraction new version
+% OPTIONS
+ % Number of frames to chunk from movie when doing ROI estimation, to reduce RAM usage.
+ movieChunks = 100;
+% Normal PCA-ICA, binary masks
+[roiSignals, ~] = ciapkg.signal_extraction.computeSignalsFromImages(pcaicaAnalysisOutput.IcaFilters,inputMoviePath,'frameList',[],'readMovieChunks',1,'threshold',0.4,'nFramesPerChunk',movieChunks,'weightSignalByImage',0);
+% Weighted PCA-ICA, trace based on weighted pixel values of eahc ROI
+[roiSignalsWeighted, ~] = ciapkg.signal_extraction.computeSignalsFromImages(pcaicaAnalysisOutput.IcaFilters,inputMoviePath,'frameList',[],'readMovieChunks',1,'threshold',0.4,'nFramesPerChunk',movieChunks,'weightSignalByImage',1);
+```
\ No newline at end of file
diff --git a/docs/docs/help_motion_correction.md b/docs/docs/help_motion_correction.md
new file mode 100644
index 0000000..9e93cc9
--- /dev/null
+++ b/docs/docs/help_motion_correction.md
@@ -0,0 +1,78 @@
+Scripts to register movies to remove motion.
+
+## Calculating final transformation after multiple registration iterations.
+It is possible to use the output from motion correction (often done with `turboregMovie` or `modelPreprocessMovieFunction`) to transform the movie at later times if needed. There are two ways to do this:
+
+- iteratively perform each motion correction step (e.g. same order as in `modelPreprocessMovieFunction`) or
+- create the translation/skew/rotation matrices for each step using `ResultsOutOriginal` and combine for all iterations as `totalTranformMatrix = (R2'*S2'*T2'*R1'*S1'*T1')'`. Note the order matters.
+ - Where `1, 2, ...` indicate matrix for iterations `1,2,...` and `R, S, T` are rotation, skew (shear + scale) and translation matrices, respectively.
+ - For 3 iterations would be `(R3'*S3'*T3'*R2'*S2'*T2'*R1'*S1'*T1')'` or alternatively `T1*S1*R1*T3*S3*R2*T3*S3*R3`.
+ - For translation/rotation matrices, use definitions in https://www.mathworks.com/help/images/matrix-representation-of-geometric-transformations.html to construct them.
+
+## Turboreg
+### Compiling `turboreg` and `transfturboreg` mex file
+* Can compile on your system using the following command
+```Matlab
+mex('-v', '-largeArrayDims','-I.', 'turboreg.c','.\BsplnTrf.c','.\BsplnWgt.c','.\convolve.c','.\getPut.c','.\main.c','.\phil.c','.\pyrFilt.c','.\pyrGetSz.c','.\quant.c','.\reg0.c','.\reg1.c','.\reg2.c','.\reg3.c','.\regFlt3d.c','.\svdcmp.c')
+
+mex('-v', '-largeArrayDims','-I.', 'transfturboreg.c','.\BsplnTrf.c','.\BsplnWgt.c','.\convolve.c','.\getPut.c','.\main.c','.\phil.c','.\pyrFilt.c','.\pyrGetSz.c','.\quant.c','.\reg0.c','.\reg1.c','.\reg2.c','.\reg3.c','.\regFlt3d.c','.\svdcmp.c')
+```
+
+ * For Linux users: http://www.walkingrandomly.com/?p=2694
+```Matlab
+mex('-v', 'GCC="/usr/bin/gcc-4.9"', '-largeArrayDims','CFLAGS="\$CFLAGS -std=c99"','-I.', 'turboreg.c','./BsplnTrf.c','./BsplnWgt.c','./convolve.c','./getPut.c','./main.c','./phil.c','./pyrFilt.c','./pyrGetSz.c','./quant.c','./reg0.c','./reg1.c','./reg2.c','./reg3.c','./regFlt3d.c','./svdcmp.c')
+
+mex('-v', 'GCC="/usr/bin/gcc-4.9"', '-largeArrayDims','CFLAGS="\$CFLAGS -std=c99"','-I.', 'transfturboreg.c','./BsplnTrf.c','./BsplnWgt.c','./convolve.c','./getPut.c','./main.c','./phil.c','./pyrFilt.c','./pyrGetSz.c','./quant.c','./reg0.c','./reg1.c','./reg2.c','./reg3.c','./regFlt3d.c','./svdcmp.c')
+```
+
+* Below is an example usage of `turboregMovie`.
+* To use imageJ in Matlab, download Fiji (
+
+-  (starting 2020.09.16)
+-  (starting 2020.09.22)
+
\ No newline at end of file
diff --git a/docs/docs/example_data.md b/docs/docs/example_data.md
new file mode 100644
index 0000000..710ad4d
--- /dev/null
+++ b/docs/docs/example_data.md
@@ -0,0 +1,7 @@
+### Example data
+
+The `{{ site.name }}` repository contains several example one-photon calcium imaging movies from miniature microscope experiments in freely moving rodents.
+
+To download example data, run `loadDependencies` module (e.g. `obj.loadDependencies`) and select `Download test one-photon data.` option to download one-photon miniature microscope example datasets to use for testing `{{ site.name }}` preprocessing, cell extraction, and cell classification code. The data will be located in the `data` folder within the repository root directory.
+
+Else run `example_downloadTestData.m` from the MATLAB command line with the working directory set to the `{{ site.name }}` repository.
\ No newline at end of file
diff --git a/docs/docs/help_animal_tracking.md b/docs/docs/help_animal_tracking.md
index b31c80b..e0fc098 100644
--- a/docs/docs/help_animal_tracking.md
+++ b/docs/docs/help_animal_tracking.md
@@ -1,28 +1,51 @@
-# Tracking
+# Animal tracking
-Code for ImageJ and Matlab based image tracking.
+Code for ImageJ- and Matlab-based image tracking.
# ImageJ based tracking
-Functions needed (have entire `miniscope_analysis` loaded anyways):
+Functions needed (have entire `{{ site.name }}` pipeline loaded anyways to ensure all dependencies are met):
- `mm_tracking.ijm` is the tracking function for use in ImageJ, place in
-`plugins` folder.
-- `removeIncorrectObjs.m` is a function to clean-up the ImageJ output.
-- `createTrackingOverlayVideo` is a way to check the output from the
-tracking by overlaying mouse tracker onto the video.
+`plugins` folder. It is found in the `ciapkg\tracking` folder of the repository.
+- `removeIncorrectObjs()` is a function to clean-up the ImageJ output.
+- `createTrackingOverlayVideo()` is a function that allows users to check the output from the tracking by overlaying the mouse tracker onto the video.
## Instructions for ImageJ and Matlab
+Run `mm_tracking` from the `Plugins` menu of ImageJ. Several settings to check:
+
+- `number of session folders to analyze` - this will indicate how many movies you want to analyze. Depending on `analyze movies from a list file` setting, a GUI will appear asking you to select movies to analyze (preferably AVIs) or a text file with a list of movies.
+- `pixel to cm conversion, distance (cm)` - Make sure pixel to cm conversion indicates a measurable distance in the video, e.g. from one side of the box to another. The program will ask you to draw this distance to estimate pixels/cm.
+- `crop stack?` - keep this selected, as allow removal of parts of movie where the animal will not go, improving results.
+- `erode and dilate` - likely keep this selected, as it essentially makes sure the mouse is a solid object and smooths the tracking.
+- `analyze movies from a list file` - select this option if you have a text file with the location of each movie to be analyzed on a new line. Use this to analyze many movies in batch.
+- `hide loaded movie` - uncheck this to be able to visualize as ImageJ completes each step in the processing. Leave checked to improve speed of analysis.
+
Example screen after running `mm_tracking` within ImageJ, click to expand.
-
-Once ImageJ is finished, within Matlab run the following code (cleans up the ImageJ tracking by removing small objects and adding NaNs for missing frames along with making a movie to check output). Modify to point toward paths specific for your data.
+
+
+
+Once ImageJ is finished, within `MATLAB` run the following code (cleans up the ImageJ tracking by removing small objects and adding NaNs for missing frames along with making a movie to check output). Modify to point toward paths specific for your data.
```Matlab
% CSV file from imageJ and AVI movie path used in ImageJ
moviePath = 'PATH_TO_AVI_USED_IN_IMAEJ';
csvPath = 'PATH_TO_CSV_OUTPUT_BY_IMAGEJ';
% clean up tracking
[trackingTableFilteredCell] = removeIncorrectObjs(csvPath,'inputMovie',{moviePath});
+```
+
+## Example output from animal in open field during miniature microscope imaging
+
+
+Tracking of an animal over time (green = early in session, red = late in session).
+
+
+## Tracking video
+
+The tracking video can be used to quickly validate that the animal is being correctly tracked.
+
+```Matlab
% make tracking video
% frames to use as example check
nFrames=1500:2500;
@@ -31,12 +54,10 @@ inputMovie = loadMovieList(moviePath,'frameList',nFrames);
playMovie(inputTrackingVideo);
```
-## Example output from 2017_09_11_p540_m381_openfield01_091112017
-
-
+Overlay of tracking (red circle) on the mouse in a specific frame.

-# Matlab based tracking
+
diff --git a/docs/docs/help_cell_extraction.md b/docs/docs/help_cell_extraction.md
new file mode 100644
index 0000000..e91bffb
--- /dev/null
+++ b/docs/docs/help_cell_extraction.md
@@ -0,0 +1,2 @@
+# Cell extraction
+
diff --git a/docs/docs/help_contrast.md b/docs/docs/help_contrast.md
new file mode 100644
index 0000000..724fbe9
--- /dev/null
+++ b/docs/docs/help_contrast.md
@@ -0,0 +1,32 @@
+# Interpreting displayed movies
+
+It may appear at times that `{{ site.name }}` has introduced noise into movies after processing. However, normally that noise is already there in the movie. In the case of one-photon miniature microscope movies that noise is often small relative to the baseline background fluorescence, hence not noticeable. However, after dF/F0 or spatial filtering users will be able to see the noise more easily as it will have a relatively greater magnitude compared to the signal now present in the movie.
+
+Further, in many cases the contrast is automatically adjusted in `{{ site.name }}` GUIs to boost likelihood users will see cells and other biologically relevant features (even though underlying matrix values are not changed), which can sometimes lead to the perception that noise is being added depending on viewing raw vs. dF/F0 or other preprocessed movies.
+
+## Example
+
+For example of what this looks like, take one of the example movies in the {{ site.name }} repository: `2014_04_01_p203_m19_check01`.
+
+### Raw movie
+
+A frame from the raw movie looks like:
+
+
+
+Applying a simple bandpass or highpass filter (to remove the low frequency background) leads to the below (keeping contrast/brightness the same as the raw movie image):
+
+
+
+However, if we adjust the contrast, we can now see some of the noise present in the higher frequency components of the raw movie that was otherwise obscured or would be hard to see in the raw movie with the high baseline present:
+
+
+
+### dF/F0
+Now compare to dF/F0 of that same raw movie without any motion correction, etc. we see the below:
+
+
+
+However, if you adjust the contrast, you get the below image, where the noise is much more pronounced:
+
+
\ No newline at end of file
diff --git a/docs/docs/help_large_movie_analysis.md b/docs/docs/help_large_movie_analysis.md
new file mode 100644
index 0000000..f619438
--- /dev/null
+++ b/docs/docs/help_large_movie_analysis.md
@@ -0,0 +1,44 @@
+# Analyzing large movies
+
+Some movies are larger than the available RAM on users analysis computer. Below are several ways that the underlying functions in `CIAPKG` can be used to analyze large movies.
+
+## Playing large movies from disk
+
+To directly and quickly visualize large or long movies from disk, directly input the movie path into `playMovie` as below.
+
+```Matlab
+% Full path to the movie
+inputMoviePath = 'path/to/movie.h5';
+
+playMovie(inputMoviePath);
+```
+
+Which will produce a GUI as below that will play the movie back.
+
+
+
+## ROI signal extraction
+The below code is an example ROI signal extraction for large movie in chunks from disk after analyzing a small chunk with PCA-ICA to obtain reference masks. Modify `inputMoviePath` to a full path to your movie (HDF5, TIF, AVI, and ISXD supported).
+
+```MATLAB
+% Full path to the movie
+inputMoviePath = 'path/to/movie.h5';
+
+%% =======PCA-ICA
+% Run PCA-ICA on only a subset of frames.
+% OPTIONS
+ % Vector of frames to analyze for PCA-ICA
+ framesToAnalyzePcaIca = 1:300;
+ % Number of PCs and ICs to request
+ nPCs = 250; nICs = 200;
+[pcaicaAnalysisOutput] = ciapkg.signal_extraction.runPcaIca(inputMoviePath,nPCs,nICs,'frameList',framesToAnalyzePcaIca,'mu',0.1,'max_iter',1e3);
+
+%% =======ROI extraction new version
+% OPTIONS
+ % Number of frames to chunk from movie when doing ROI estimation, to reduce RAM usage.
+ movieChunks = 100;
+% Normal PCA-ICA, binary masks
+[roiSignals, ~] = ciapkg.signal_extraction.computeSignalsFromImages(pcaicaAnalysisOutput.IcaFilters,inputMoviePath,'frameList',[],'readMovieChunks',1,'threshold',0.4,'nFramesPerChunk',movieChunks,'weightSignalByImage',0);
+% Weighted PCA-ICA, trace based on weighted pixel values of eahc ROI
+[roiSignalsWeighted, ~] = ciapkg.signal_extraction.computeSignalsFromImages(pcaicaAnalysisOutput.IcaFilters,inputMoviePath,'frameList',[],'readMovieChunks',1,'threshold',0.4,'nFramesPerChunk',movieChunks,'weightSignalByImage',1);
+```
\ No newline at end of file
diff --git a/docs/docs/help_motion_correction.md b/docs/docs/help_motion_correction.md
new file mode 100644
index 0000000..9e93cc9
--- /dev/null
+++ b/docs/docs/help_motion_correction.md
@@ -0,0 +1,78 @@
+Scripts to register movies to remove motion.
+
+## Calculating final transformation after multiple registration iterations.
+It is possible to use the output from motion correction (often done with `turboregMovie` or `modelPreprocessMovieFunction`) to transform the movie at later times if needed. There are two ways to do this:
+
+- iteratively perform each motion correction step (e.g. same order as in `modelPreprocessMovieFunction`) or
+- create the translation/skew/rotation matrices for each step using `ResultsOutOriginal` and combine for all iterations as `totalTranformMatrix = (R2'*S2'*T2'*R1'*S1'*T1')'`. Note the order matters.
+ - Where `1, 2, ...` indicate matrix for iterations `1,2,...` and `R, S, T` are rotation, skew (shear + scale) and translation matrices, respectively.
+ - For 3 iterations would be `(R3'*S3'*T3'*R2'*S2'*T2'*R1'*S1'*T1')'` or alternatively `T1*S1*R1*T3*S3*R2*T3*S3*R3`.
+ - For translation/rotation matrices, use definitions in https://www.mathworks.com/help/images/matrix-representation-of-geometric-transformations.html to construct them.
+
+## Turboreg
+### Compiling `turboreg` and `transfturboreg` mex file
+* Can compile on your system using the following command
+```Matlab
+mex('-v', '-largeArrayDims','-I.', 'turboreg.c','.\BsplnTrf.c','.\BsplnWgt.c','.\convolve.c','.\getPut.c','.\main.c','.\phil.c','.\pyrFilt.c','.\pyrGetSz.c','.\quant.c','.\reg0.c','.\reg1.c','.\reg2.c','.\reg3.c','.\regFlt3d.c','.\svdcmp.c')
+
+mex('-v', '-largeArrayDims','-I.', 'transfturboreg.c','.\BsplnTrf.c','.\BsplnWgt.c','.\convolve.c','.\getPut.c','.\main.c','.\phil.c','.\pyrFilt.c','.\pyrGetSz.c','.\quant.c','.\reg0.c','.\reg1.c','.\reg2.c','.\reg3.c','.\regFlt3d.c','.\svdcmp.c')
+```
+
+ * For Linux users: http://www.walkingrandomly.com/?p=2694
+```Matlab
+mex('-v', 'GCC="/usr/bin/gcc-4.9"', '-largeArrayDims','CFLAGS="\$CFLAGS -std=c99"','-I.', 'turboreg.c','./BsplnTrf.c','./BsplnWgt.c','./convolve.c','./getPut.c','./main.c','./phil.c','./pyrFilt.c','./pyrGetSz.c','./quant.c','./reg0.c','./reg1.c','./reg2.c','./reg3.c','./regFlt3d.c','./svdcmp.c')
+
+mex('-v', 'GCC="/usr/bin/gcc-4.9"', '-largeArrayDims','CFLAGS="\$CFLAGS -std=c99"','-I.', 'transfturboreg.c','./BsplnTrf.c','./BsplnWgt.c','./convolve.c','./getPut.c','./main.c','./phil.c','./pyrFilt.c','./pyrGetSz.c','./quant.c','./reg0.c','./reg1.c','./reg2.c','./reg3.c','./regFlt3d.c','./svdcmp.c')
+```
+
+* Below is an example usage of `turboregMovie`.
+* To use imageJ in Matlab, download Fiji ( @@ -14,14 +16,25 @@ Created by __Biafra Ahanonu, PhD__.
`{{ site.name }}` (pronounced cheetah, formerly `calciumImagingAnalysis` [`CIAPKG`]) is a software package for analysis of one- and two-photon calcium imaging datasets.
-__Download the software at [https://github.com/bahanonu/calciumImagingAnalysis](https://github.com/bahanonu/calciumImagingAnalysis).__
+__Download the software at [https://github.com/bahanonu/ciatah](https://github.com/bahanonu/ciatah).__
@@ -14,14 +16,25 @@ Created by __Biafra Ahanonu, PhD__.
`{{ site.name }}` (pronounced cheetah, formerly `calciumImagingAnalysis` [`CIAPKG`]) is a software package for analysis of one- and two-photon calcium imaging datasets.
-__Download the software at [https://github.com/bahanonu/calciumImagingAnalysis](https://github.com/bahanonu/calciumImagingAnalysis).__
+__Download the software at [https://github.com/bahanonu/ciatah](https://github.com/bahanonu/ciatah).__
 - This user guide contains instructions to setup, run, and troubleshoot `{{ site.name }}`.
- Note: `{{ site.name }}` is a class (`{{ site.namelow }}` within MATLAB) with various GUIs to allow processing of calcium imaging data. In addition, users can access the underlying `{{ site.name }}` functions to make custom workflows. See [Custom command-line pipelines](api_example_pipeline.md).
+
+## Guides
+
+Below are recordings for users who want to learn more about calcium imaging analysis.
+
- Read my overview of calcium imaging analysis methods at [Calcium imaging cell identification and fluorescence activity trace reconstruction, part 1](https://bahanonu.com/brain/#c20181209).
+### Webinar
+This webinar gives an overview of calcium imaging analysis (with a focus on CIAtah) along with tips for improving experiments and analysis: https://info.inscopix.com/inscopix-inspire-view-webinarbiafra-ahanonu-signal-in-the-noise-distinguishing-relevant-neural-activity-in-calcium-imaging.
+
+### Workshop tutorial
+This recording gives an overview of setting up and using CIAtah: https://www.youtube.com/watch?v=I6abW3uuJJw.
+
@@ -54,12 +67,19 @@ __Download the software at [https://github.com/bahanonu/calciumImagingAnalysis](
`{{ site.name }}` features:
-- Includes a GUI to allow users to do large-scale batch analysis, accessed via the repository's `{{ site.name }}` class.
-- The underlying functions can also be used to create GUI-less, command line-ready analysis pipelines. Functions located in `ciapkg` and `+ciapkg` sub-folders.
-- Includes all major calcium imaging analysis steps: pre-processing (motion correction, spatiotemporal downsampling, spatial filtering, relative fluorescence calculation, etc.), support for multiple cell-extraction methods, automated cell classification (coming soon!), cross-session cell alignment, and more.
-- Has several example one- and two-photon calcium imaging datasets that it will automatically download to help users test out the package.
-- Includes code for determining animal position (e.g. in open-field assay).
-- Supports [Neurodata Without Borders](https://www.nwb.org/) data standard (see [calcium imaging tutorial](https://neurodatawithoutborders.github.io/matnwb/tutorials/html/ophys.html)) for reading/writing cell-extraction (e.g. outputs of PCA-ICA, CELLMax, CNMF, CNMF-E, etc.). Supports reading and writing NWB movie files with continued integration with NWB planned.
+- A GUI with different modules for large-scale batch analysis.
+- `CIAtah` functions (in `ciapkg`/`+ciapkg` folders) can be used to create GUI-less, command line-ready analysis pipelines.
+- Includes all major calcium imaging analysis steps:
+ - movie visualization (including reading from disk, for fast viewing of large movies)
+ - pre-processing (motion correction, spatiotemporal downsampling, spatial filtering, relative fluorescence calculation, etc.)
+ - support for multiple cell-extraction methods (CELLMax, PCA-ICA, CNMF, CNMF-E, EXTRACT, etc.)
+ - manual classification of cells via GUIs,
+ - automated cell classification (i.e. CLEAN algorithm, coming soon!),
+ - cross-session cell alignment, and more.
+- Includes example one- and two-photon calcium imaging datasets for testing `ciatah`.
+- Supports a plethora of major imaging movie file formats: HDF5, NWB, AVI, ISXD [Inscopix], TIFF, and [Bio-Formats](https://www.openmicroscopy.org/bio-formats/) compatible formats (Olympus [OIR] and Zeiss [CZI and LSM] currently, additional support to be added or upon request).
+- Supports [Neurodata Without Borders](https://www.nwb.org/) data standard (see [calcium imaging tutorial](https://neurodatawithoutborders.github.io/matnwb/tutorials/html/ophys.html)) for reading/writing cell-extraction and imaging movie files.
+- Animal position tracking (e.g. in open-field assay).
- Requires `MATLAB`.

diff --git a/docs/docs/install.md b/docs/docs/install.md
index 66c5236..adfee5d 100644
--- a/docs/docs/install.md
+++ b/docs/docs/install.md
@@ -1,12 +1,28 @@
-## Quick start guide
+# Quick start guide
Below are steps needed to quickly get started using the `{{ site.name }}` software package in MATLAB.
-- Clone the `{{ site.name }}` repository (using [GitHub desktop](https://desktop.github.com/) or command line) or download the repository zip and unzip.
+## Install
+- Clone the `{{ site.name }}` repository (using [GitHub desktop](https://desktop.github.com/) or command line) or download the repository zip and unzip (e.g. run below MATLAB command).
- Point the MATLAB path to the `{{ site.name }}` root folder (*NOT* `@{{ code.mainclass }}` sub-folder in the repository).
- Alternatively, download the package from `File Exchange` using the Add-Ons explorer in MATLAB. See `{{ site.name }}` entry at:
[](https://www.mathworks.com/matlabcentral/fileexchange/75466-calciumimaginganalysis) or https://www.mathworks.com/matlabcentral/fileexchange/75466-calciumimaginganalysis.
-- Run the below MATLAB commands.
+
+ ```Matlab
+ % Optional: this will set MATLAB working folder to the default user path. Make sure you have read/write permissions.
+ try; cd(userpath); catch; end;
+
+ % Download and unzip current repository
+ unzip('https://github.com/bahanonu/{{ code.mainclass }}/archive/master.zip');
+
+ % Make CIAtah the working folder
+ cd('{{ code.mainclass }}-master')
+ ```
+
+
+## Setup `{{ site.name }}`
+
+- Run `{{ site.name }}` using the below MATLAB commands. Call `obj;` in the MATLAB command window each time you want to go back to the main GUI. __Note: `calciumImagingAnalysis` class is now called `ciatah`, all functionality is the same.__
```MATLAB
% Run these commands in MATLAB to get started.
@@ -25,7 +41,7 @@ obj % then hit enter, no semicolon!
- [Optional] Users on Windows systems should download `Everything` (https://www.voidtools.com/). It is a very useful and extremely fast search engine for files and folders on a computer that can allow users to quickly get lists of folders then need to analyze in `{{ site.name }}`.
- [Optional] Users who want to run analysis via the command line can run `edit ciapkg.demo.cmdLinePipeline` and run each segment of code there to see what commands are needed to perform each step. It assumes you have already run `example_downloadTestData`.
-### `{{ site.name }}` main GUI notes
+## `{{ site.name }}` main GUI notes
- All main decisions for choosing a method/procedure to run, cell-extraction algorithm, and which folders to analyze are in a single window.
- The GUI will real-time update the selected folders based on the selections in the subject, assay, and folder filter areas.
- Sections not relevant for a specific method are grayed out.
@@ -42,7 +58,7 @@ __Certain sections become available when user selects the appropriate method (e.
- This user guide contains instructions to setup, run, and troubleshoot `{{ site.name }}`.
- Note: `{{ site.name }}` is a class (`{{ site.namelow }}` within MATLAB) with various GUIs to allow processing of calcium imaging data. In addition, users can access the underlying `{{ site.name }}` functions to make custom workflows. See [Custom command-line pipelines](api_example_pipeline.md).
+
+## Guides
+
+Below are recordings for users who want to learn more about calcium imaging analysis.
+
- Read my overview of calcium imaging analysis methods at [Calcium imaging cell identification and fluorescence activity trace reconstruction, part 1](https://bahanonu.com/brain/#c20181209).
+### Webinar
+This webinar gives an overview of calcium imaging analysis (with a focus on CIAtah) along with tips for improving experiments and analysis: https://info.inscopix.com/inscopix-inspire-view-webinarbiafra-ahanonu-signal-in-the-noise-distinguishing-relevant-neural-activity-in-calcium-imaging.
+
+### Workshop tutorial
+This recording gives an overview of setting up and using CIAtah: https://www.youtube.com/watch?v=I6abW3uuJJw.
+
@@ -54,12 +67,19 @@ __Download the software at [https://github.com/bahanonu/calciumImagingAnalysis](
`{{ site.name }}` features:
-- Includes a GUI to allow users to do large-scale batch analysis, accessed via the repository's `{{ site.name }}` class.
-- The underlying functions can also be used to create GUI-less, command line-ready analysis pipelines. Functions located in `ciapkg` and `+ciapkg` sub-folders.
-- Includes all major calcium imaging analysis steps: pre-processing (motion correction, spatiotemporal downsampling, spatial filtering, relative fluorescence calculation, etc.), support for multiple cell-extraction methods, automated cell classification (coming soon!), cross-session cell alignment, and more.
-- Has several example one- and two-photon calcium imaging datasets that it will automatically download to help users test out the package.
-- Includes code for determining animal position (e.g. in open-field assay).
-- Supports [Neurodata Without Borders](https://www.nwb.org/) data standard (see [calcium imaging tutorial](https://neurodatawithoutborders.github.io/matnwb/tutorials/html/ophys.html)) for reading/writing cell-extraction (e.g. outputs of PCA-ICA, CELLMax, CNMF, CNMF-E, etc.). Supports reading and writing NWB movie files with continued integration with NWB planned.
+- A GUI with different modules for large-scale batch analysis.
+- `CIAtah` functions (in `ciapkg`/`+ciapkg` folders) can be used to create GUI-less, command line-ready analysis pipelines.
+- Includes all major calcium imaging analysis steps:
+ - movie visualization (including reading from disk, for fast viewing of large movies)
+ - pre-processing (motion correction, spatiotemporal downsampling, spatial filtering, relative fluorescence calculation, etc.)
+ - support for multiple cell-extraction methods (CELLMax, PCA-ICA, CNMF, CNMF-E, EXTRACT, etc.)
+ - manual classification of cells via GUIs,
+ - automated cell classification (i.e. CLEAN algorithm, coming soon!),
+ - cross-session cell alignment, and more.
+- Includes example one- and two-photon calcium imaging datasets for testing `ciatah`.
+- Supports a plethora of major imaging movie file formats: HDF5, NWB, AVI, ISXD [Inscopix], TIFF, and [Bio-Formats](https://www.openmicroscopy.org/bio-formats/) compatible formats (Olympus [OIR] and Zeiss [CZI and LSM] currently, additional support to be added or upon request).
+- Supports [Neurodata Without Borders](https://www.nwb.org/) data standard (see [calcium imaging tutorial](https://neurodatawithoutborders.github.io/matnwb/tutorials/html/ophys.html)) for reading/writing cell-extraction and imaging movie files.
+- Animal position tracking (e.g. in open-field assay).
- Requires `MATLAB`.

diff --git a/docs/docs/install.md b/docs/docs/install.md
index 66c5236..adfee5d 100644
--- a/docs/docs/install.md
+++ b/docs/docs/install.md
@@ -1,12 +1,28 @@
-## Quick start guide
+# Quick start guide
Below are steps needed to quickly get started using the `{{ site.name }}` software package in MATLAB.
-- Clone the `{{ site.name }}` repository (using [GitHub desktop](https://desktop.github.com/) or command line) or download the repository zip and unzip.
+## Install
+- Clone the `{{ site.name }}` repository (using [GitHub desktop](https://desktop.github.com/) or command line) or download the repository zip and unzip (e.g. run below MATLAB command).
- Point the MATLAB path to the `{{ site.name }}` root folder (*NOT* `@{{ code.mainclass }}` sub-folder in the repository).
- Alternatively, download the package from `File Exchange` using the Add-Ons explorer in MATLAB. See `{{ site.name }}` entry at:
[](https://www.mathworks.com/matlabcentral/fileexchange/75466-calciumimaginganalysis) or https://www.mathworks.com/matlabcentral/fileexchange/75466-calciumimaginganalysis.
-- Run the below MATLAB commands.
+
+ ```Matlab
+ % Optional: this will set MATLAB working folder to the default user path. Make sure you have read/write permissions.
+ try; cd(userpath); catch; end;
+
+ % Download and unzip current repository
+ unzip('https://github.com/bahanonu/{{ code.mainclass }}/archive/master.zip');
+
+ % Make CIAtah the working folder
+ cd('{{ code.mainclass }}-master')
+ ```
+
+
+## Setup `{{ site.name }}`
+
+- Run `{{ site.name }}` using the below MATLAB commands. Call `obj;` in the MATLAB command window each time you want to go back to the main GUI. __Note: `calciumImagingAnalysis` class is now called `ciatah`, all functionality is the same.__
```MATLAB
% Run these commands in MATLAB to get started.
@@ -25,7 +41,7 @@ obj % then hit enter, no semicolon!
- [Optional] Users on Windows systems should download `Everything` (https://www.voidtools.com/). It is a very useful and extremely fast search engine for files and folders on a computer that can allow users to quickly get lists of folders then need to analyze in `{{ site.name }}`.
- [Optional] Users who want to run analysis via the command line can run `edit ciapkg.demo.cmdLinePipeline` and run each segment of code there to see what commands are needed to perform each step. It assumes you have already run `example_downloadTestData`.
-### `{{ site.name }}` main GUI notes
+## `{{ site.name }}` main GUI notes
- All main decisions for choosing a method/procedure to run, cell-extraction algorithm, and which folders to analyze are in a single window.
- The GUI will real-time update the selected folders based on the selections in the subject, assay, and folder filter areas.
- Sections not relevant for a specific method are grayed out.
@@ -42,7 +58,7 @@ __Certain sections become available when user selects the appropriate method (e.
 -### Additional quick start notes
+## Additional quick start notes
- See additional details in [Processing calcium imaging data](#processing-calcium-imaging-data) for running the full processing pipeline.
- Settings used to pre-process imaging movies (`modelPreprocessMovie` module) are stored inside the HDF5 file to allow `{{ site.name }}` to load them again later.
diff --git a/docs/docs/install_alt.md b/docs/docs/install_alt.md
new file mode 100644
index 0000000..9aebdac
--- /dev/null
+++ b/docs/docs/install_alt.md
@@ -0,0 +1,19 @@
+# Installation
+
+Note, this is an alternative method of installation to that outlined in [Quick Start](install.md).
+
+Clone the `{{ site.name }}` repository or download the repository zip and unzip.
+
+- Point the MATLAB path to the `{{ site.name }}` folder.
+- Run `loadBatchFxns.m` before using functions in the directory. This adds all needed directories and sub-directories to the MATLAB path.
+- Type `obj = {{ code.mainclass }};` into MATLAB command window and follow instructions that appear after to add data and run analysis.
+- Run the `{{ code.mainclass }}` class method `loadDependencies` or type `obj.loadDependencies` after initializing a `{{ code.mainclass }}` object into the command window to download and add Fiji to path, download CNMF/CNMF-E repositories, download/setup CVX (for CNMF/CNMF-E), and download example data.
+
+Note
+
+- Place `{{ site.name }}` in a folder where MATLAB will have write permissions, as it also creates a `private` subdirectory to store some user information along with downloading required external software packages.
+- `file_exchange` folder contains File Exchange functions used by `{{ site.name }}`.
+- In general, it is best to set the MATLAB startup directory to the `{{ site.name }}` folder. This allows `java.opts` and `startup.m` to set the correct Java memory requirements and load the correct folders into the MATLAB path.
+- If `{{ site.name }}` IS NOT the startup folder, place `java.opts` wherever the MATLAB startup folder is so the correct Java memory requirements are set (important for using ImageJ/Miji in MATLAB).
+- If it appears an old `{{ site.name }}` repository is loaded after pulling a new version, run `restoredefaultpath` and check that old `{{ site.name }}` folders are not in the MATLAB path.
+
\ No newline at end of file
diff --git a/docs/docs/install_gui_notes.md b/docs/docs/install_gui_notes.md
new file mode 100644
index 0000000..f9e1c0b
--- /dev/null
+++ b/docs/docs/install_gui_notes.md
@@ -0,0 +1,40 @@
+# Additional quick start notes
+
+- See additional details on the [Processing calcium imaging data](https://bahanonu.github.io/ciatah/pipeline_overview/) page for running the full processing pipeline.
+- Settings used to pre-process imaging movies (`modelPreprocessMovie` module) are stored inside the processed HDF5 movie to allow `CIAtah` to load them again later.
+- To force load all directories, including most external software packages (in `_external_programs` folder), type `ciapkg.loadAllDirs;` into MATLAB command line. This is most relevant when you need to access specific functions in an outside repository that are normally hidden until needed.
+- When issues are encountered, first check the [Common issues and fixes](https://bahanonu.github.io/ciatah/help_issues/) page to see if a solution is there. Else, submit a new issue or email Biafra.
+- There are two sets of test data that are downloaded:
+ - __Single session analysis__: `data\2014_04_01_p203_m19_check01_raw` can be used to test the pipeline until the cross-session alignment step.
+ - __Batch analysis__: `data\batch` contains three imaging sessions that should be processed and can then be used for the cross-session alignment step. Users should try these sessions to get used to batched analysis.
+- For Fiji dependency, when path to `Miji.m` (e.g. `\Fiji.app\scripts` folder) is requested, likely in `[CIAtah directory]\_external_programs\FIJI_FOLDER\Fiji.app\scripts` where `FIJI_FOLDER` varies depending on OS, unless the user requested a custom path or on OSX (in which case, find Fiji the install directory).
+ - If you run into Java heap space memory errors when Miji tries to load Fiji in MATLAB, make sure "java.opts" file is in MATLAB start-up folder or that the `CIAtah` root folder is the MATLAB start-up folder ([instructions on changing](https://www.mathworks.com/help/matlab/matlab_env/matlab-startup-folder.html)).
+- `CIAtah` often uses [regular expressions](https://www.cheatography.com/davechild/cheat-sheets/regular-expressions/) to find relevant movie and other files in folders to analyze.
+ - For example, by default it looks for any movie files in a folder containing `concat`, e.g. `concat_recording_20140401_180333.h5` (test data). If you have a file called `rawData_2019_01_01_myInterestingExperiment.avi` and all your raw data files start with `rawData_` then change the regular expression to `rawData_` when requested by the repository. See `setMovieInfo` module to change after adding new folders.
+- `CIAtah` generally assumes users have imaging data associated with *one* imaging session and animal in a given folder. Follow folder naming conventions in [Data formats](https://bahanonu.github.io/ciatah/data/#preferred-folder-naming-format) for the best experience.
+- External software packages are downloaded into `_external_programs` folder and should be placed there if done manually.
+
+Users can alternatively run setup as below.
+```MATLAB
+% Run these commands in MATLAB to get started.
+
+% Loads all directories
+loadBatchFxns;
+
+% Loads the class into an object for use in this session
+obj = ciatah;
+
+% Download and load dependent software packages into "_external_programs" folder.
+% Also download test data into "data" folder.
+% Normally only need to one once after first downloading CIAtah package.
+obj.loadDependencies;
+
+% Add folders containing imaging data.
+obj.modelAddNewFolders;
+
+% [optional] Set the names CIAtah will look for in each folder
+obj.setMovieInfo;
+
+% Open class menu to pick module to run.
+obj.runPipeline; % then hit enter!
+```
diff --git a/docs/docs/pipeline_detailed_all.md b/docs/docs/pipeline_detailed_all.md
new file mode 100644
index 0000000..0a9b667
--- /dev/null
+++ b/docs/docs/pipeline_detailed_all.md
@@ -0,0 +1,20 @@
+# Detailed {{ site.name }} processing pipeline
+
+The following detailed pipeline assumes you have started a {{ site.name }} object using the below command:
+
+```Matlab
+obj = ciatah;
+```
+
+This is the one-page version of the guide. Visit the individual pages by using the sidebar at left, can be easier to follow for some.
+
+{!pipeline_detailed_downsample_raw.md!}
+{!pipeline_detailed_preprocess_check.md!}
+{!pipeline_detailed_preprocess.md!}
+{!pipeline_detailed_modify_movies.md!}
+{!pipeline_detailed_signal_extraction.md!}
+{!pipeline_detailed_signal_extraction_load.md!}
+{!pipeline_detailed_signal_extraction_validation.md!}
+{!pipeline_detailed_signal_sorting_manual.md!}
+{!pipeline_detailed_signal_region_analysis.md!}
+{!pipeline_detailed_cross_session.md!}
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_cross_session.md b/docs/docs/pipeline_detailed_cross_session.md
new file mode 100644
index 0000000..e0fc8a8
--- /dev/null
+++ b/docs/docs/pipeline_detailed_cross_session.md
@@ -0,0 +1,33 @@
+---
+title: Cross-session cell alignment.
+---
+
+# Cross-session cell alignment with `computeMatchObjBtwnTrials`
+
+This step allows users to align cells across imaging sessions (e.g. those taken on different days). See the `Cross session cell alignment` wiki page for more details and notes on cross-session alignment.
+
+- Users run `computeMatchObjBtwnTrials` to do cross-day alignment (first row in pictures below).
+- Users then run `viewMatchObjBtwnSessions` to get a sense for how well the alignment ran.
+- `computeCellDistances` and `computeCrossDayDistancesAlignment` allow users to compute the within session pairwise Euclidean centroid distance for all cells and the cross-session pairwise distance for all global matched cells, respectively.
+
+
+
+Users can then get the matrix that gives the session IDs
+
+```Matlab
+% Global IDs is a matrix of [globalID sessionID]
+% Each (globalID, sessionID) pair gives the within session ID for that particular global ID
+globalIDs = alignmentStruct.globalIDs;
+
+```
+
+## View cross-session cell alignment with `viewMatchObjBtwnSessions`
+
+To evaluate how well cross-session alignment works, `computeMatchObjBtwnTrials` will automatically run `viewMatchObjBtwnSessions` at the end, but users can also run it separately after alignment. The left are raw dorsal striatum cell maps from a single animal. The right shows after cross-session alignment; color is used to indicate a global ID cell (e.g. the same cell matched across multiple days). Thus, same color cell = same cell across sessions.
+
+
-### Additional quick start notes
+## Additional quick start notes
- See additional details in [Processing calcium imaging data](#processing-calcium-imaging-data) for running the full processing pipeline.
- Settings used to pre-process imaging movies (`modelPreprocessMovie` module) are stored inside the HDF5 file to allow `{{ site.name }}` to load them again later.
diff --git a/docs/docs/install_alt.md b/docs/docs/install_alt.md
new file mode 100644
index 0000000..9aebdac
--- /dev/null
+++ b/docs/docs/install_alt.md
@@ -0,0 +1,19 @@
+# Installation
+
+Note, this is an alternative method of installation to that outlined in [Quick Start](install.md).
+
+Clone the `{{ site.name }}` repository or download the repository zip and unzip.
+
+- Point the MATLAB path to the `{{ site.name }}` folder.
+- Run `loadBatchFxns.m` before using functions in the directory. This adds all needed directories and sub-directories to the MATLAB path.
+- Type `obj = {{ code.mainclass }};` into MATLAB command window and follow instructions that appear after to add data and run analysis.
+- Run the `{{ code.mainclass }}` class method `loadDependencies` or type `obj.loadDependencies` after initializing a `{{ code.mainclass }}` object into the command window to download and add Fiji to path, download CNMF/CNMF-E repositories, download/setup CVX (for CNMF/CNMF-E), and download example data.
+
+Note
+
+- Place `{{ site.name }}` in a folder where MATLAB will have write permissions, as it also creates a `private` subdirectory to store some user information along with downloading required external software packages.
+- `file_exchange` folder contains File Exchange functions used by `{{ site.name }}`.
+- In general, it is best to set the MATLAB startup directory to the `{{ site.name }}` folder. This allows `java.opts` and `startup.m` to set the correct Java memory requirements and load the correct folders into the MATLAB path.
+- If `{{ site.name }}` IS NOT the startup folder, place `java.opts` wherever the MATLAB startup folder is so the correct Java memory requirements are set (important for using ImageJ/Miji in MATLAB).
+- If it appears an old `{{ site.name }}` repository is loaded after pulling a new version, run `restoredefaultpath` and check that old `{{ site.name }}` folders are not in the MATLAB path.
+
\ No newline at end of file
diff --git a/docs/docs/install_gui_notes.md b/docs/docs/install_gui_notes.md
new file mode 100644
index 0000000..f9e1c0b
--- /dev/null
+++ b/docs/docs/install_gui_notes.md
@@ -0,0 +1,40 @@
+# Additional quick start notes
+
+- See additional details on the [Processing calcium imaging data](https://bahanonu.github.io/ciatah/pipeline_overview/) page for running the full processing pipeline.
+- Settings used to pre-process imaging movies (`modelPreprocessMovie` module) are stored inside the processed HDF5 movie to allow `CIAtah` to load them again later.
+- To force load all directories, including most external software packages (in `_external_programs` folder), type `ciapkg.loadAllDirs;` into MATLAB command line. This is most relevant when you need to access specific functions in an outside repository that are normally hidden until needed.
+- When issues are encountered, first check the [Common issues and fixes](https://bahanonu.github.io/ciatah/help_issues/) page to see if a solution is there. Else, submit a new issue or email Biafra.
+- There are two sets of test data that are downloaded:
+ - __Single session analysis__: `data\2014_04_01_p203_m19_check01_raw` can be used to test the pipeline until the cross-session alignment step.
+ - __Batch analysis__: `data\batch` contains three imaging sessions that should be processed and can then be used for the cross-session alignment step. Users should try these sessions to get used to batched analysis.
+- For Fiji dependency, when path to `Miji.m` (e.g. `\Fiji.app\scripts` folder) is requested, likely in `[CIAtah directory]\_external_programs\FIJI_FOLDER\Fiji.app\scripts` where `FIJI_FOLDER` varies depending on OS, unless the user requested a custom path or on OSX (in which case, find Fiji the install directory).
+ - If you run into Java heap space memory errors when Miji tries to load Fiji in MATLAB, make sure "java.opts" file is in MATLAB start-up folder or that the `CIAtah` root folder is the MATLAB start-up folder ([instructions on changing](https://www.mathworks.com/help/matlab/matlab_env/matlab-startup-folder.html)).
+- `CIAtah` often uses [regular expressions](https://www.cheatography.com/davechild/cheat-sheets/regular-expressions/) to find relevant movie and other files in folders to analyze.
+ - For example, by default it looks for any movie files in a folder containing `concat`, e.g. `concat_recording_20140401_180333.h5` (test data). If you have a file called `rawData_2019_01_01_myInterestingExperiment.avi` and all your raw data files start with `rawData_` then change the regular expression to `rawData_` when requested by the repository. See `setMovieInfo` module to change after adding new folders.
+- `CIAtah` generally assumes users have imaging data associated with *one* imaging session and animal in a given folder. Follow folder naming conventions in [Data formats](https://bahanonu.github.io/ciatah/data/#preferred-folder-naming-format) for the best experience.
+- External software packages are downloaded into `_external_programs` folder and should be placed there if done manually.
+
+Users can alternatively run setup as below.
+```MATLAB
+% Run these commands in MATLAB to get started.
+
+% Loads all directories
+loadBatchFxns;
+
+% Loads the class into an object for use in this session
+obj = ciatah;
+
+% Download and load dependent software packages into "_external_programs" folder.
+% Also download test data into "data" folder.
+% Normally only need to one once after first downloading CIAtah package.
+obj.loadDependencies;
+
+% Add folders containing imaging data.
+obj.modelAddNewFolders;
+
+% [optional] Set the names CIAtah will look for in each folder
+obj.setMovieInfo;
+
+% Open class menu to pick module to run.
+obj.runPipeline; % then hit enter!
+```
diff --git a/docs/docs/pipeline_detailed_all.md b/docs/docs/pipeline_detailed_all.md
new file mode 100644
index 0000000..0a9b667
--- /dev/null
+++ b/docs/docs/pipeline_detailed_all.md
@@ -0,0 +1,20 @@
+# Detailed {{ site.name }} processing pipeline
+
+The following detailed pipeline assumes you have started a {{ site.name }} object using the below command:
+
+```Matlab
+obj = ciatah;
+```
+
+This is the one-page version of the guide. Visit the individual pages by using the sidebar at left, can be easier to follow for some.
+
+{!pipeline_detailed_downsample_raw.md!}
+{!pipeline_detailed_preprocess_check.md!}
+{!pipeline_detailed_preprocess.md!}
+{!pipeline_detailed_modify_movies.md!}
+{!pipeline_detailed_signal_extraction.md!}
+{!pipeline_detailed_signal_extraction_load.md!}
+{!pipeline_detailed_signal_extraction_validation.md!}
+{!pipeline_detailed_signal_sorting_manual.md!}
+{!pipeline_detailed_signal_region_analysis.md!}
+{!pipeline_detailed_cross_session.md!}
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_cross_session.md b/docs/docs/pipeline_detailed_cross_session.md
new file mode 100644
index 0000000..e0fc8a8
--- /dev/null
+++ b/docs/docs/pipeline_detailed_cross_session.md
@@ -0,0 +1,33 @@
+---
+title: Cross-session cell alignment.
+---
+
+# Cross-session cell alignment with `computeMatchObjBtwnTrials`
+
+This step allows users to align cells across imaging sessions (e.g. those taken on different days). See the `Cross session cell alignment` wiki page for more details and notes on cross-session alignment.
+
+- Users run `computeMatchObjBtwnTrials` to do cross-day alignment (first row in pictures below).
+- Users then run `viewMatchObjBtwnSessions` to get a sense for how well the alignment ran.
+- `computeCellDistances` and `computeCrossDayDistancesAlignment` allow users to compute the within session pairwise Euclidean centroid distance for all cells and the cross-session pairwise distance for all global matched cells, respectively.
+
+
+
+Users can then get the matrix that gives the session IDs
+
+```Matlab
+% Global IDs is a matrix of [globalID sessionID]
+% Each (globalID, sessionID) pair gives the within session ID for that particular global ID
+globalIDs = alignmentStruct.globalIDs;
+
+```
+
+## View cross-session cell alignment with `viewMatchObjBtwnSessions`
+
+To evaluate how well cross-session alignment works, `computeMatchObjBtwnTrials` will automatically run `viewMatchObjBtwnSessions` at the end, but users can also run it separately after alignment. The left are raw dorsal striatum cell maps from a single animal. The right shows after cross-session alignment; color is used to indicate a global ID cell (e.g. the same cell matched across multiple days). Thus, same color cell = same cell across sessions.
+
+ +
+ +
+## Save cross-session cell alignment with `modelSaveMatchObjBtwnTrials`
+
+Users can save out the alignment structure by running `modelSaveMatchObjBtwnTrials`. This will allow users to select a folder where `{{ site.name }}` will save a MAT-file with the alignment structure information for each animal.
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_downsample_raw.md b/docs/docs/pipeline_detailed_downsample_raw.md
new file mode 100644
index 0000000..0b1a049
--- /dev/null
+++ b/docs/docs/pipeline_detailed_downsample_raw.md
@@ -0,0 +1,30 @@
+---
+title: Spatially downsample raw movies or convert to HDF5
+---
+
+# Spatially downsample raw movies or convert to HDF5 with `modelDownsampleRawMovies`
+
+Users have the ability to spatially downsample raw movies, often necessary to denoise the data, save storage space, and improve runtimes of later processing steps. For most data, users can downsample 2 or 4 times in each spatial dimension while still retaining sufficient pixels per cell to facilitate cell-extraction.
+
+To run, either select `modelDownsampleRawMovies` in the GUI menu or type the below command after initializing a {{ site.name }} obj.
+
+```Matlab
+obj.modelDownsampleRawMovies;
+```
+
+This will pop-up the following screen. Users can
+- input several folders where ISXD files are by separating each folder path with a comma (`Folder(s) where raw HDF5s are located`),
+- specify a common root folder to save files to (`Folder to save downsampled HDF5s to:`),
+- and input a root directory that contains the sub-folders with the raw data (`Decompression source root folder(s)`).
+The function will automatically put each file in its corresponding folder, __make sure folder names are unique__ (this should be done anyways for data analysis reasons).
+
+
+
+
+## Converting Inscopix ISXD files to HDF5
+
+To convert from Inscopix ISXD file format (output by nVista v3+ and nVoke) to HDF5 run `modelDownsampleRawMovies` without changing the regular expression or make sure it looks for `.*.isxd` or similar. Users will need the latest version of the [Inscopix Data Processing Software](https://www.inscopix.com/nVista#Data_Analysis) as these functions take advantage of their API. If {{ site.name }} cannot automatically find the API, it will ask the user to direct it to the _root_ location of the Inscopix Data Processing Software (see below).
+
+
+
+
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_modify_movies.md b/docs/docs/pipeline_detailed_modify_movies.md
new file mode 100644
index 0000000..beaba21
--- /dev/null
+++ b/docs/docs/pipeline_detailed_modify_movies.md
@@ -0,0 +1,13 @@
+---
+title: Manual movie cropping.
+---
+
+# Manual movie cropping with `modelModifyMovies`
+
+If users need to eliminate specific regions of their movie before running cell extraction, that option is provided. Users select a region using an ImageJ interface and select `done` when they want to move onto the next movie or start the cropping. Movies have `NaNs` or `0s` added in the cropped region rather than changing the dimensions of the movie.
+
+This is generally advised for movies such as miniature microscope movies imaged through a GRIN lens probe where the outside or edge or the GRIN lens are visible. This can lead to large fluctuations that can throw off some algorithms (e.g. PCA-ICA can end up assigning many components to these "signals").
+
+
+
+
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_preprocess.md b/docs/docs/pipeline_detailed_preprocess.md
new file mode 100644
index 0000000..bb6b2e4
--- /dev/null
+++ b/docs/docs/pipeline_detailed_preprocess.md
@@ -0,0 +1,40 @@
+---
+title: Preprocessing calcium imaging movies
+---
+
+# Preprocessing calcium imaging movies with `modelPreprocessMovie`
+
+After users instantiate an object of the `{{ site.name }}` class and enter a folder, they can start preprocessing of their calcium imaging data with `modelPreprocessMovie`.
+
+- See below for a series of windows to get started, the options for motion correction, cropping unneeded regions, Δ_F/F_, and temporal downsampling were selected for use in the study associated with this repository.
+- If users have not specified the path to Miji, a window appears asking them to select the path to Miji's `scripts` folder.
+- If users are using the test dataset, it is recommended that they do not use temporal downsampling.
+- Vertical and horizontal stripes in movies (e.g. CMOS camera artifacts) can be removed via `stripeRemoval` step. Remember to select correct `stripOrientationRemove`,`stripSize`, and `stripfreqLowExclude` options in the preprocessing options menu.
+
+
+
+Next the user is presented with a series of options for motion correction, image registration, and cropping.:
+
+- The options highlighted in green are those that should be considered by users.
+- Users can over their mouse over each option to get tips on what they mean.
+- In particular, make sure that `inputDatasetName` is correct for HDF5 files and that `fileFilterRegexp` matches the form of the calcium imaging movie files to be analyzed.
+- After this, the user is asked to let the algorithm know how many frames of the movie to analyze (defaults to all frames).
+- Then the user is asked to select a region to use for motion correction. In general, it is best to select areas with high contrast and static markers such as blood vessels. Stay away from the edge of the movie or areas outside the brain (e.g. the edge of microendoscope GRIN lens in one-photon miniature microscope movies).
+
+
+
+## Save/load preprocessing settings
+
+Users can also enable saving and loading of previously selected pre-processing settings by changing the red option below.
+
+
+
+Settings loaded from previous run (e.g. of `modelPreprocessMovie`) or file (e.g. from `viewMovieRegistrationTest` runs) are highlighted in orange. Settings that user has just changed are still highlighted in green.
+
+
+
+The algorithm will then run all the requested preprocessing steps and presented the user with the option of viewing a slice of the processed file. Users have now completed pre-processing.
+
+
+
+
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_preprocess_check.md b/docs/docs/pipeline_detailed_preprocess_check.md
new file mode 100644
index 0000000..b2408a7
--- /dev/null
+++ b/docs/docs/pipeline_detailed_preprocess_check.md
@@ -0,0 +1,20 @@
+---
+title: Check movie registration before pre-processing
+---
+
+# Check movie registration before pre-processing with `viewMovieRegistrationTest`
+
+Users should spatially filter one-photon or other data with background noise (e.g. neuropil). To get a feel for how the different spatial filtering affects SNR/movie data before running the full processing pipeline, run `viewMovieRegistrationTest` module. Then select either `matlab divide by lowpass before registering` or `matlab bandpass before registering` then change `filterBeforeRegFreqLow` and `filterBeforeRegFreqHigh` settings, see below.
+
+Within each folder will be a sub-folder called `preprocRunTest` inside of which is a series of sub-folders called `preprocRun##` that will contain a file called `settings.mat` that can be loaded into `modelPreprocessMovie` so the same settings that worked during the test can be used during the actual pre-processing run.
+
+
+
+- You'll get an output like the below:
+ - __A__: The top left is without any filtering while the other 3 are with different bandpass filtering options.
+ - __B__: Cell ΔF/F intensity profile from the raw movie. Obtain by selecting `Analyze->Plot profile` from Fiji menu after selecting a square segment running through a cell.
+ - __C__: Same cell ΔF/F intensity profile from the bottom/left movie (note the y-axis is the same as above). Obtained in same manner as __B__.
+
+
+
+
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_signal_extraction.md b/docs/docs/pipeline_detailed_signal_extraction.md
new file mode 100644
index 0000000..c7e4fef
--- /dev/null
+++ b/docs/docs/pipeline_detailed_signal_extraction.md
@@ -0,0 +1,21 @@
+---
+title: Automated cell extraction.
+---
+
+# Extracting cells with `modelExtractSignalsFromMovie`
+
+Users can run PCA-ICA, EXTRACT, CNMF, CNMF-E, and ROI cell extraction by following the below set of option screens. Details on running the new Schnitzer lab cell-extraction methods (e.g. CELLMax) will be added here after they are released.
+
+We normally estimate the number of PCs and ICs on the high end, manually sort to get an estimate of the number of cells, then run PCA-ICA again with IC 1.5-3x the number of cells and PCs 1-1.5x number of ICs.
+
+To run CNMF or CNMF-E, run `loadDependencies` module (e.g. `obj.loadDependencies`) after {{ site.name }} class is loaded. CVX (a CNMF dependency) will also be downloaded and `cvx_setup` run to automatically set it up.
+
+
+
+The resulting output (on _Figure 45+_) at the end should look something like:
+
+
+
+
+
+
diff --git a/docs/docs/pipeline_detailed_signal_extraction_load.md b/docs/docs/pipeline_detailed_signal_extraction_load.md
new file mode 100644
index 0000000..dc63639
--- /dev/null
+++ b/docs/docs/pipeline_detailed_signal_extraction_load.md
@@ -0,0 +1,34 @@
+---
+title: Loading cell extraction data.
+---
+
+# Loading cell-extraction output data for custom scripts
+
+Users can load outputs from cell extraction using the below command. This will then allow users to use the images and activity traces for downstream analysis as needed.
+
+```Matlab
+[inputImages,inputSignals,infoStruct,algorithmStr,inputSignals2] = ciapkg.io.loadSignalExtraction('pathToFile');
+```
+
+Note, the outputs correspond to the below:
+
+- `inputImages` - 3D or 4D matrix containing cells and their spatial information, format: [x y nCells].
+- `inputSignals` - 2D matrix containing activity traces in [nCells nFrames] format.
+- `infoStruct` - contains information about the file, e.g. the 'description' property that can contain information about the algorithm.
+- `algorithmStr` - String of the algorithm name.
+- `inputSignals2` - same as inputSignals but for secondary traces an algorithm outputs.
+
+
+
+## Loading cell-extraction output data with `modelVarsFromFiles`
+
+In general, after running cell-extraction (`modelExtractSignalsFromMovie`) on a dataset, run the `modelVarsFromFiles` module. This allows `{{ site.name }}` to load/pre-load information about that cell-extraction run.
+
+If you had to restart MATLAB or are just loading {{ site.name }} fresh but have previously run cell extraction, run this method before doing anything else with that cell-extraction data.
+
+A menu will pop-up like below when `modelVarsFromFiles` is loaded, you can normally just leave the defaults as is.
+
+
+
+
+
diff --git a/docs/docs/pipeline_detailed_signal_extraction_validation.md b/docs/docs/pipeline_detailed_signal_extraction_validation.md
new file mode 100644
index 0000000..4d69977
--- /dev/null
+++ b/docs/docs/pipeline_detailed_signal_extraction_validation.md
@@ -0,0 +1,13 @@
+---
+title: Validating cell extraction data.
+---
+
+# Validating cell extraction with `viewCellExtractionOnMovie`
+
+After users have run cell extraction, they should check that cells are not being missed during the process. Running the method `viewCellExtractionOnMovie` will create a movie with outlines of cell extraction outputs overlaid on the movie.
+
+Below is an example, with black outlines indicating location of cell extraction outputs. If users see active cells (red flashes) that are not outlined, that indicates either exclusion or other parameters should be altered in the previous `modelExtractSignalsFromMovie` cell extraction step.
+
+
+
+
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_signal_region_analysis.md b/docs/docs/pipeline_detailed_signal_region_analysis.md
new file mode 100644
index 0000000..f56140a
--- /dev/null
+++ b/docs/docs/pipeline_detailed_signal_region_analysis.md
@@ -0,0 +1,11 @@
+---
+title: Removing cells not within region of interest.
+---
+
+# Removing cells not within brain region with `modelModifyRegionAnalysis`
+
+If the imaging field-of-view includes cells from other brain regions, they can be removed using `modelModifyRegionAnalysis`
+
+
+
+
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_signal_sorting_manual.md b/docs/docs/pipeline_detailed_signal_sorting_manual.md
new file mode 100644
index 0000000..539e506
--- /dev/null
+++ b/docs/docs/pipeline_detailed_signal_sorting_manual.md
@@ -0,0 +1,73 @@
+---
+title: Sorting cell extraction outputs.
+---
+
+# Sorting cell extraction outputs with `computeManualSortSignals`
+
+
+
+## Save cross-session cell alignment with `modelSaveMatchObjBtwnTrials`
+
+Users can save out the alignment structure by running `modelSaveMatchObjBtwnTrials`. This will allow users to select a folder where `{{ site.name }}` will save a MAT-file with the alignment structure information for each animal.
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_downsample_raw.md b/docs/docs/pipeline_detailed_downsample_raw.md
new file mode 100644
index 0000000..0b1a049
--- /dev/null
+++ b/docs/docs/pipeline_detailed_downsample_raw.md
@@ -0,0 +1,30 @@
+---
+title: Spatially downsample raw movies or convert to HDF5
+---
+
+# Spatially downsample raw movies or convert to HDF5 with `modelDownsampleRawMovies`
+
+Users have the ability to spatially downsample raw movies, often necessary to denoise the data, save storage space, and improve runtimes of later processing steps. For most data, users can downsample 2 or 4 times in each spatial dimension while still retaining sufficient pixels per cell to facilitate cell-extraction.
+
+To run, either select `modelDownsampleRawMovies` in the GUI menu or type the below command after initializing a {{ site.name }} obj.
+
+```Matlab
+obj.modelDownsampleRawMovies;
+```
+
+This will pop-up the following screen. Users can
+- input several folders where ISXD files are by separating each folder path with a comma (`Folder(s) where raw HDF5s are located`),
+- specify a common root folder to save files to (`Folder to save downsampled HDF5s to:`),
+- and input a root directory that contains the sub-folders with the raw data (`Decompression source root folder(s)`).
+The function will automatically put each file in its corresponding folder, __make sure folder names are unique__ (this should be done anyways for data analysis reasons).
+
+
+
+
+## Converting Inscopix ISXD files to HDF5
+
+To convert from Inscopix ISXD file format (output by nVista v3+ and nVoke) to HDF5 run `modelDownsampleRawMovies` without changing the regular expression or make sure it looks for `.*.isxd` or similar. Users will need the latest version of the [Inscopix Data Processing Software](https://www.inscopix.com/nVista#Data_Analysis) as these functions take advantage of their API. If {{ site.name }} cannot automatically find the API, it will ask the user to direct it to the _root_ location of the Inscopix Data Processing Software (see below).
+
+
+
+
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_modify_movies.md b/docs/docs/pipeline_detailed_modify_movies.md
new file mode 100644
index 0000000..beaba21
--- /dev/null
+++ b/docs/docs/pipeline_detailed_modify_movies.md
@@ -0,0 +1,13 @@
+---
+title: Manual movie cropping.
+---
+
+# Manual movie cropping with `modelModifyMovies`
+
+If users need to eliminate specific regions of their movie before running cell extraction, that option is provided. Users select a region using an ImageJ interface and select `done` when they want to move onto the next movie or start the cropping. Movies have `NaNs` or `0s` added in the cropped region rather than changing the dimensions of the movie.
+
+This is generally advised for movies such as miniature microscope movies imaged through a GRIN lens probe where the outside or edge or the GRIN lens are visible. This can lead to large fluctuations that can throw off some algorithms (e.g. PCA-ICA can end up assigning many components to these "signals").
+
+
+
+
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_preprocess.md b/docs/docs/pipeline_detailed_preprocess.md
new file mode 100644
index 0000000..bb6b2e4
--- /dev/null
+++ b/docs/docs/pipeline_detailed_preprocess.md
@@ -0,0 +1,40 @@
+---
+title: Preprocessing calcium imaging movies
+---
+
+# Preprocessing calcium imaging movies with `modelPreprocessMovie`
+
+After users instantiate an object of the `{{ site.name }}` class and enter a folder, they can start preprocessing of their calcium imaging data with `modelPreprocessMovie`.
+
+- See below for a series of windows to get started, the options for motion correction, cropping unneeded regions, Δ_F/F_, and temporal downsampling were selected for use in the study associated with this repository.
+- If users have not specified the path to Miji, a window appears asking them to select the path to Miji's `scripts` folder.
+- If users are using the test dataset, it is recommended that they do not use temporal downsampling.
+- Vertical and horizontal stripes in movies (e.g. CMOS camera artifacts) can be removed via `stripeRemoval` step. Remember to select correct `stripOrientationRemove`,`stripSize`, and `stripfreqLowExclude` options in the preprocessing options menu.
+
+
+
+Next the user is presented with a series of options for motion correction, image registration, and cropping.:
+
+- The options highlighted in green are those that should be considered by users.
+- Users can over their mouse over each option to get tips on what they mean.
+- In particular, make sure that `inputDatasetName` is correct for HDF5 files and that `fileFilterRegexp` matches the form of the calcium imaging movie files to be analyzed.
+- After this, the user is asked to let the algorithm know how many frames of the movie to analyze (defaults to all frames).
+- Then the user is asked to select a region to use for motion correction. In general, it is best to select areas with high contrast and static markers such as blood vessels. Stay away from the edge of the movie or areas outside the brain (e.g. the edge of microendoscope GRIN lens in one-photon miniature microscope movies).
+
+
+
+## Save/load preprocessing settings
+
+Users can also enable saving and loading of previously selected pre-processing settings by changing the red option below.
+
+
+
+Settings loaded from previous run (e.g. of `modelPreprocessMovie`) or file (e.g. from `viewMovieRegistrationTest` runs) are highlighted in orange. Settings that user has just changed are still highlighted in green.
+
+
+
+The algorithm will then run all the requested preprocessing steps and presented the user with the option of viewing a slice of the processed file. Users have now completed pre-processing.
+
+
+
+
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_preprocess_check.md b/docs/docs/pipeline_detailed_preprocess_check.md
new file mode 100644
index 0000000..b2408a7
--- /dev/null
+++ b/docs/docs/pipeline_detailed_preprocess_check.md
@@ -0,0 +1,20 @@
+---
+title: Check movie registration before pre-processing
+---
+
+# Check movie registration before pre-processing with `viewMovieRegistrationTest`
+
+Users should spatially filter one-photon or other data with background noise (e.g. neuropil). To get a feel for how the different spatial filtering affects SNR/movie data before running the full processing pipeline, run `viewMovieRegistrationTest` module. Then select either `matlab divide by lowpass before registering` or `matlab bandpass before registering` then change `filterBeforeRegFreqLow` and `filterBeforeRegFreqHigh` settings, see below.
+
+Within each folder will be a sub-folder called `preprocRunTest` inside of which is a series of sub-folders called `preprocRun##` that will contain a file called `settings.mat` that can be loaded into `modelPreprocessMovie` so the same settings that worked during the test can be used during the actual pre-processing run.
+
+
+
+- You'll get an output like the below:
+ - __A__: The top left is without any filtering while the other 3 are with different bandpass filtering options.
+ - __B__: Cell ΔF/F intensity profile from the raw movie. Obtain by selecting `Analyze->Plot profile` from Fiji menu after selecting a square segment running through a cell.
+ - __C__: Same cell ΔF/F intensity profile from the bottom/left movie (note the y-axis is the same as above). Obtained in same manner as __B__.
+
+
+
+
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_signal_extraction.md b/docs/docs/pipeline_detailed_signal_extraction.md
new file mode 100644
index 0000000..c7e4fef
--- /dev/null
+++ b/docs/docs/pipeline_detailed_signal_extraction.md
@@ -0,0 +1,21 @@
+---
+title: Automated cell extraction.
+---
+
+# Extracting cells with `modelExtractSignalsFromMovie`
+
+Users can run PCA-ICA, EXTRACT, CNMF, CNMF-E, and ROI cell extraction by following the below set of option screens. Details on running the new Schnitzer lab cell-extraction methods (e.g. CELLMax) will be added here after they are released.
+
+We normally estimate the number of PCs and ICs on the high end, manually sort to get an estimate of the number of cells, then run PCA-ICA again with IC 1.5-3x the number of cells and PCs 1-1.5x number of ICs.
+
+To run CNMF or CNMF-E, run `loadDependencies` module (e.g. `obj.loadDependencies`) after {{ site.name }} class is loaded. CVX (a CNMF dependency) will also be downloaded and `cvx_setup` run to automatically set it up.
+
+
+
+The resulting output (on _Figure 45+_) at the end should look something like:
+
+
+
+
+
+
diff --git a/docs/docs/pipeline_detailed_signal_extraction_load.md b/docs/docs/pipeline_detailed_signal_extraction_load.md
new file mode 100644
index 0000000..dc63639
--- /dev/null
+++ b/docs/docs/pipeline_detailed_signal_extraction_load.md
@@ -0,0 +1,34 @@
+---
+title: Loading cell extraction data.
+---
+
+# Loading cell-extraction output data for custom scripts
+
+Users can load outputs from cell extraction using the below command. This will then allow users to use the images and activity traces for downstream analysis as needed.
+
+```Matlab
+[inputImages,inputSignals,infoStruct,algorithmStr,inputSignals2] = ciapkg.io.loadSignalExtraction('pathToFile');
+```
+
+Note, the outputs correspond to the below:
+
+- `inputImages` - 3D or 4D matrix containing cells and their spatial information, format: [x y nCells].
+- `inputSignals` - 2D matrix containing activity traces in [nCells nFrames] format.
+- `infoStruct` - contains information about the file, e.g. the 'description' property that can contain information about the algorithm.
+- `algorithmStr` - String of the algorithm name.
+- `inputSignals2` - same as inputSignals but for secondary traces an algorithm outputs.
+
+
+
+## Loading cell-extraction output data with `modelVarsFromFiles`
+
+In general, after running cell-extraction (`modelExtractSignalsFromMovie`) on a dataset, run the `modelVarsFromFiles` module. This allows `{{ site.name }}` to load/pre-load information about that cell-extraction run.
+
+If you had to restart MATLAB or are just loading {{ site.name }} fresh but have previously run cell extraction, run this method before doing anything else with that cell-extraction data.
+
+A menu will pop-up like below when `modelVarsFromFiles` is loaded, you can normally just leave the defaults as is.
+
+
+
+
+
diff --git a/docs/docs/pipeline_detailed_signal_extraction_validation.md b/docs/docs/pipeline_detailed_signal_extraction_validation.md
new file mode 100644
index 0000000..4d69977
--- /dev/null
+++ b/docs/docs/pipeline_detailed_signal_extraction_validation.md
@@ -0,0 +1,13 @@
+---
+title: Validating cell extraction data.
+---
+
+# Validating cell extraction with `viewCellExtractionOnMovie`
+
+After users have run cell extraction, they should check that cells are not being missed during the process. Running the method `viewCellExtractionOnMovie` will create a movie with outlines of cell extraction outputs overlaid on the movie.
+
+Below is an example, with black outlines indicating location of cell extraction outputs. If users see active cells (red flashes) that are not outlined, that indicates either exclusion or other parameters should be altered in the previous `modelExtractSignalsFromMovie` cell extraction step.
+
+
+
+
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_signal_region_analysis.md b/docs/docs/pipeline_detailed_signal_region_analysis.md
new file mode 100644
index 0000000..f56140a
--- /dev/null
+++ b/docs/docs/pipeline_detailed_signal_region_analysis.md
@@ -0,0 +1,11 @@
+---
+title: Removing cells not within region of interest.
+---
+
+# Removing cells not within brain region with `modelModifyRegionAnalysis`
+
+If the imaging field-of-view includes cells from other brain regions, they can be removed using `modelModifyRegionAnalysis`
+
+
+
+
\ No newline at end of file
diff --git a/docs/docs/pipeline_detailed_signal_sorting_manual.md b/docs/docs/pipeline_detailed_signal_sorting_manual.md
new file mode 100644
index 0000000..539e506
--- /dev/null
+++ b/docs/docs/pipeline_detailed_signal_sorting_manual.md
@@ -0,0 +1,73 @@
+---
+title: Sorting cell extraction outputs.
+---
+
+# Sorting cell extraction outputs with `computeManualSortSignals`
+
++ {{ site.name }} cell sorting GUI +
+
+
+  +
+
+
+
 +
+
\ No newline at end of file
diff --git a/docs/docs/pipeline_overview.md b/docs/docs/pipeline_overview.md
index 70bcc41..bb2676a 100644
--- a/docs/docs/pipeline_overview.md
+++ b/docs/docs/pipeline_overview.md
@@ -5,6 +5,17 @@ The general pipeline for processing calcium imaging data is below. This reposito

+## Guides
+Below are recordings for users who want to learn more about calcium imaging analysis.
+
+### Webinar
+This webinar gives an overview of calcium imaging analysis (with a focus on CIAtah) along with tips for improving experiments and analysis: https://info.inscopix.com/inscopix-inspire-view-webinarbiafra-ahanonu-signal-in-the-noise-distinguishing-relevant-neural-activity-in-calcium-imaging.
+
+### Workshop tutorial
+This recording gives an overview of setting up and using CIAtah: https://www.youtube.com/watch?v=I6abW3uuJJw.
+
+## Workflow
+
To start using the `{{ site.name }}` software package, enter the following into the MATLAB command window.
```Matlab
diff --git a/docs/docs/questions.md b/docs/docs/questions.md
index 722f152..eace4cf 100644
--- a/docs/docs/questions.md
+++ b/docs/docs/questions.md
@@ -1,2 +1,2 @@
-## Questions?
+# Questions?
Please [open an issue on GitHub](https://github.com/bahanonu/calciumImagingAnalysis/issues) or email any additional questions not covered in the repository to `bahanonu [at] alum.mit.edu`.
\ No newline at end of file
diff --git a/docs/docs/references.md b/docs/docs/references.md
index e355133..34f51d4 100644
--- a/docs/docs/references.md
+++ b/docs/docs/references.md
@@ -1,4 +1,4 @@
-## References
+# References
Please cite [Corder*, Ahanonu*, et al. 2019](http://science.sciencemag.org/content/363/6424/276.full) _Science_ publication or the [Ahanonu, 2018](https://doi.org/10.5281/zenodo.2222294) _Zenodo_ release if you used the software package or code from this repository to advance/help your research:
diff --git a/docs/make_docs.py b/docs/make_docs.py
index 12fceab..e0414f7 100644
--- a/docs/make_docs.py
+++ b/docs/make_docs.py
@@ -4,9 +4,12 @@
# Changelog
# 2020.09.28 [22:35:51] - Added ssl module.
# 2020.09.28 [22:42:51] - Added support to automatically install missing packages. Disabled as user should do this.
+ # 2021.04.05 [16:12:12] - Since all_docs uses includes, yaml extra variables are not evaluated. This is a quick way to replace them.
import subprocess
import sys
+from bs4 import BeautifulSoup
+import re
# import socket
# import ssl
@@ -20,10 +23,39 @@
# pip install mdx_truly_sane_lists
# pip install pymdown-extensions
# pip install mkdocs-macros-plugin
+# pip install mkdocs-markdownextradata-plugin
+# pip install markdown-include
# If you want to view the docs before building
# mkdocs serve
# Build the documents
# mkdocs build
-subprocess.check_call([sys.executable, "-m", "mkdocs", "build"])
\ No newline at end of file
+subprocess.check_call([sys.executable, "-m", "mkdocs", "build"])
+
+
+# Since all_docs uses includes, yaml extra variables are not evaluated. This is a quick way to replace them.
+f = open('site/all_docs/index.html','r')
+html = f.read()
+f.close()
+soup = BeautifulSoup(html)
+
+extraDict = {
+ '{{ site.name }}': 'CIAtah',
+ '{{ site.namelow }}': 'ciatah',
+ '{{ site.nameold }}': 'calciumImagingAnalysis',
+ '{{ code.mainclass }}': 'ciatah',
+ '{{ code.mainclass }}': 'ciatah'
+}
+for key in extraDict:
+ target = soup.find_all(text=re.compile(re.escape(key)))
+ # target = soup.find_all(text=re.compile(r'{{ site.name }}'))
+ for v in target:
+ v.replace_with(v.replace(key,extraDict[key]))
+
+# print soup
+f = open('site/all_docs/index.html','w')
+soup_string = str(soup)
+soup_string = soup_string.replace('{{ code.mainclass }}', 'ciatah')
+f.write(soup_string)
+f.close()
\ No newline at end of file
diff --git a/docs/mkdocs.yml b/docs/mkdocs.yml
index effd537..e039b1e 100644
--- a/docs/mkdocs.yml
+++ b/docs/mkdocs.yml
@@ -1,4 +1,5 @@
site_name: CIAtah
+site_author: Biafra Ahanonu
# For online, to end in /data instead of /data.html
use_directory_urls: true
theme:
@@ -21,6 +22,8 @@ theme:
- navigation.expand
plugins:
- macros
+ # - search
+ # - markdownextradata: {}
markdown_extensions:
- toc:
permalink: true
@@ -30,6 +33,14 @@ markdown_extensions:
- pymdownx.highlight
- pymdownx.superfences
- pymdownx.inlinehilite
+ - pymdownx.snippets:
+ base_path: docs
+ - markdown_include.include:
+ base_path: docs
+ # headingOffset: 0
+ headingOffset: 1
+ # inheritHeadingDepth: True
+ # inheritHeadingDepth: False
- pymdownx.highlight:
linenums: true
linenums_style: pymdownx-inline
@@ -56,11 +67,14 @@ extra:
nameold: calciumImagingAnalysis
code:
mainclass: ciatah
+ package: ciapkg
# NAVIGATION
nav:
- Home: index.md
- - One-page readme: alldocs.md
+ - End-to-end:
+ # - One-page readme: alldocs.md
+ - One-page readme: all_docs.md
- Setup:
- Quick Start: install.md
- Install: install_alt.md
@@ -70,23 +84,35 @@ nav:
- Data formats: data.md
- Notes: notes.md
- Organization: organization.md
- - Processing data:
- - Overview: pipeline_overview.md
- - Step-by-step: pipeline_detailed.md
- - Animal tracking: pipeline_animal_tracking.md
+ - Processing imaging data:
+ - Overview: pipeline_overview.md
+ # - Step-by-step: pipeline_detailed.md
+ - One-page step-by-step: pipeline_detailed_all.md
+ - 1| Downsample raw movies: pipeline_detailed_downsample_raw.md
+ - 2| Check pre-process settings: pipeline_detailed_preprocess_check.md
+ - 3| Processing imaging movies: pipeline_detailed_preprocess.md
+ - 4| Movie clean-up: pipeline_detailed_modify_movies.md
+ - 5| Cell extraction: pipeline_detailed_signal_extraction.md
+ - 6| Loading cell extraction outputs: pipeline_detailed_signal_extraction_load.md
+ - 7| Cell extraction validation: pipeline_detailed_signal_extraction_validation.md
+ - 8| Manual cell sorting: pipeline_detailed_signal_sorting_manual.md
+ - 9| Region-specific analysis: pipeline_detailed_signal_region_analysis.md
+ - 10| Cross-session alignment: pipeline_detailed_cross_session.md
+ - Animal tracking:
+ - Animal tracking (ImageJ+MATLAB): pipeline_animal_tracking.md
- API:
- Custom pipelines: api_example_pipeline.md
- ciapkg: api_ciapkg.md
- Help:
- Issues and fixes: help_issues.md
- -
+
+
\ No newline at end of file
diff --git a/docs/docs/pipeline_overview.md b/docs/docs/pipeline_overview.md
index 70bcc41..bb2676a 100644
--- a/docs/docs/pipeline_overview.md
+++ b/docs/docs/pipeline_overview.md
@@ -5,6 +5,17 @@ The general pipeline for processing calcium imaging data is below. This reposito

+## Guides
+Below are recordings for users who want to learn more about calcium imaging analysis.
+
+### Webinar
+This webinar gives an overview of calcium imaging analysis (with a focus on CIAtah) along with tips for improving experiments and analysis: https://info.inscopix.com/inscopix-inspire-view-webinarbiafra-ahanonu-signal-in-the-noise-distinguishing-relevant-neural-activity-in-calcium-imaging.
+
+### Workshop tutorial
+This recording gives an overview of setting up and using CIAtah: https://www.youtube.com/watch?v=I6abW3uuJJw.
+
+## Workflow
+
To start using the `{{ site.name }}` software package, enter the following into the MATLAB command window.
```Matlab
diff --git a/docs/docs/questions.md b/docs/docs/questions.md
index 722f152..eace4cf 100644
--- a/docs/docs/questions.md
+++ b/docs/docs/questions.md
@@ -1,2 +1,2 @@
-## Questions?
+# Questions?
Please [open an issue on GitHub](https://github.com/bahanonu/calciumImagingAnalysis/issues) or email any additional questions not covered in the repository to `bahanonu [at] alum.mit.edu`.
\ No newline at end of file
diff --git a/docs/docs/references.md b/docs/docs/references.md
index e355133..34f51d4 100644
--- a/docs/docs/references.md
+++ b/docs/docs/references.md
@@ -1,4 +1,4 @@
-## References
+# References
Please cite [Corder*, Ahanonu*, et al. 2019](http://science.sciencemag.org/content/363/6424/276.full) _Science_ publication or the [Ahanonu, 2018](https://doi.org/10.5281/zenodo.2222294) _Zenodo_ release if you used the software package or code from this repository to advance/help your research:
diff --git a/docs/make_docs.py b/docs/make_docs.py
index 12fceab..e0414f7 100644
--- a/docs/make_docs.py
+++ b/docs/make_docs.py
@@ -4,9 +4,12 @@
# Changelog
# 2020.09.28 [22:35:51] - Added ssl module.
# 2020.09.28 [22:42:51] - Added support to automatically install missing packages. Disabled as user should do this.
+ # 2021.04.05 [16:12:12] - Since all_docs uses includes, yaml extra variables are not evaluated. This is a quick way to replace them.
import subprocess
import sys
+from bs4 import BeautifulSoup
+import re
# import socket
# import ssl
@@ -20,10 +23,39 @@
# pip install mdx_truly_sane_lists
# pip install pymdown-extensions
# pip install mkdocs-macros-plugin
+# pip install mkdocs-markdownextradata-plugin
+# pip install markdown-include
# If you want to view the docs before building
# mkdocs serve
# Build the documents
# mkdocs build
-subprocess.check_call([sys.executable, "-m", "mkdocs", "build"])
\ No newline at end of file
+subprocess.check_call([sys.executable, "-m", "mkdocs", "build"])
+
+
+# Since all_docs uses includes, yaml extra variables are not evaluated. This is a quick way to replace them.
+f = open('site/all_docs/index.html','r')
+html = f.read()
+f.close()
+soup = BeautifulSoup(html)
+
+extraDict = {
+ '{{ site.name }}': 'CIAtah',
+ '{{ site.namelow }}': 'ciatah',
+ '{{ site.nameold }}': 'calciumImagingAnalysis',
+ '{{ code.mainclass }}': 'ciatah',
+ '{{ code.mainclass }}': 'ciatah'
+}
+for key in extraDict:
+ target = soup.find_all(text=re.compile(re.escape(key)))
+ # target = soup.find_all(text=re.compile(r'{{ site.name }}'))
+ for v in target:
+ v.replace_with(v.replace(key,extraDict[key]))
+
+# print soup
+f = open('site/all_docs/index.html','w')
+soup_string = str(soup)
+soup_string = soup_string.replace('{{ code.mainclass }}', 'ciatah')
+f.write(soup_string)
+f.close()
\ No newline at end of file
diff --git a/docs/mkdocs.yml b/docs/mkdocs.yml
index effd537..e039b1e 100644
--- a/docs/mkdocs.yml
+++ b/docs/mkdocs.yml
@@ -1,4 +1,5 @@
site_name: CIAtah
+site_author: Biafra Ahanonu
# For online, to end in /data instead of /data.html
use_directory_urls: true
theme:
@@ -21,6 +22,8 @@ theme:
- navigation.expand
plugins:
- macros
+ # - search
+ # - markdownextradata: {}
markdown_extensions:
- toc:
permalink: true
@@ -30,6 +33,14 @@ markdown_extensions:
- pymdownx.highlight
- pymdownx.superfences
- pymdownx.inlinehilite
+ - pymdownx.snippets:
+ base_path: docs
+ - markdown_include.include:
+ base_path: docs
+ # headingOffset: 0
+ headingOffset: 1
+ # inheritHeadingDepth: True
+ # inheritHeadingDepth: False
- pymdownx.highlight:
linenums: true
linenums_style: pymdownx-inline
@@ -56,11 +67,14 @@ extra:
nameold: calciumImagingAnalysis
code:
mainclass: ciatah
+ package: ciapkg
# NAVIGATION
nav:
- Home: index.md
- - One-page readme: alldocs.md
+ - End-to-end:
+ # - One-page readme: alldocs.md
+ - One-page readme: all_docs.md
- Setup:
- Quick Start: install.md
- Install: install_alt.md
@@ -70,23 +84,35 @@ nav:
- Data formats: data.md
- Notes: notes.md
- Organization: organization.md
- - Processing data:
- - Overview: pipeline_overview.md
- - Step-by-step: pipeline_detailed.md
- - Animal tracking: pipeline_animal_tracking.md
+ - Processing imaging data:
+ - Overview: pipeline_overview.md
+ # - Step-by-step: pipeline_detailed.md
+ - One-page step-by-step: pipeline_detailed_all.md
+ - 1| Downsample raw movies: pipeline_detailed_downsample_raw.md
+ - 2| Check pre-process settings: pipeline_detailed_preprocess_check.md
+ - 3| Processing imaging movies: pipeline_detailed_preprocess.md
+ - 4| Movie clean-up: pipeline_detailed_modify_movies.md
+ - 5| Cell extraction: pipeline_detailed_signal_extraction.md
+ - 6| Loading cell extraction outputs: pipeline_detailed_signal_extraction_load.md
+ - 7| Cell extraction validation: pipeline_detailed_signal_extraction_validation.md
+ - 8| Manual cell sorting: pipeline_detailed_signal_sorting_manual.md
+ - 9| Region-specific analysis: pipeline_detailed_signal_region_analysis.md
+ - 10| Cross-session alignment: pipeline_detailed_cross_session.md
+ - Animal tracking:
+ - Animal tracking (ImageJ+MATLAB): pipeline_animal_tracking.md
- API:
- Custom pipelines: api_example_pipeline.md
- ciapkg: api_ciapkg.md
- Help:
- Issues and fixes: help_issues.md
- - Data
: index.md
+ - Data
: blank.md
- Inscopix: help_inscopix.md
- Neurodata Without Borders: help_nwb.md
- - Interface
: index.md
+ - Interface
: blank.md
- Movie display and noise: help_contrast.md
- - Analysis
: index.md
+ - Analysis
: blank.md
- Analysis methods: help_analysis_methods.md
- Large movie analysis: help_large_movie_analysis.md
- Stripe removal: help_stripe_removal.md
diff --git a/loadBatchFxns.m b/loadBatchFxns.m
index 24408e7..975d8c4 100644
--- a/loadBatchFxns.m
+++ b/loadBatchFxns.m
@@ -21,6 +21,7 @@ function loadBatchFxns(varargin)
% 2020.06.05 [23:35:43] - If user doesn't have Parallel Toolbox, still works
% 2020.07.21 [14:11:42] - Fix to make sure all sub-folders (not just the root) are also removed in the case of certain external_programs.
% 2021.02.01 [15:19:40] - Update `_external_programs` to call ciapkg.getDirExternalPrograms() to standardize call across all functions.
+ % 2021.06.20 [00:22:38] - Added manageMiji('startStop','closeAllWindows'); support.
% TODO
%
@@ -126,6 +127,16 @@ function loadBatchFxns(varargin)
display(repmat('@',1,7))
end
+ if workerCheck==1
+ else
+ if loadMijiCheck==0
+ disp('Skipping loading of Miji.')
+ else
+ manageMiji('startStop','setupImageJ');
+ end
+ end
+ loadMijiCheck = 0;
+
if loadMijiCheck==0
disp('Skipping loading of Miji.')
elseif workerCheck==1