diff --git a/+ciapkg/+io/loadDependencies.m b/+ciapkg/+io/loadDependencies.m
index da38ca3..a8cfb01 100644
--- a/+ciapkg/+io/loadDependencies.m
+++ b/+ciapkg/+io/loadDependencies.m
@@ -15,6 +15,7 @@ function loadDependencies(varargin)
% 2021.01.22 [13:42:36] - NWB from specific release to reduce compatibility errors.
% 2021.02.01 [15:10:41] - Separated into non-class function for use in more functions without needing to load CIAtah class.
% 2021.02.01 [15:19:40] - Update `_external_programs` to call ciapkg.getDirExternalPrograms() to standardize call across all functions.
+ % 2021.03.20 [18:12:20] - Added EXTRACT support to list of functions to download.
% TODO
% Verify all dependencies download and if not ask user to download again.
@@ -22,11 +23,11 @@ function loadDependencies(varargin)
% DESCRIPTION
options.externalProgramsDir = ciapkg.getDirExternalPrograms();
options.guiEnabled = 1;
- options.dependencyStr = {'downloadMiji','downloadCnmfGithubRepositories','example_downloadTestData','loadMiji','downloadNeuroDataWithoutBorders'};
+ options.dependencyStr = {'downloadMiji','downloadCnmfGithubRepositories','example_downloadTestData','loadMiji','downloadNeuroDataWithoutBorders','downloadEXTRACT'};
- options.dispStr = {'Download Fiji (to run Miji)','Download CNMF, CNMF-E, and CVX code.','Download test one- and two photon datasets.','Load Fiji/Miji into MATLAB path.','Download NWB (NeuroDataWithoutBorders)'};
+ options.dispStr = {'Download Fiji (to run Miji)','Download CNMF, CNMF-E, and CVX code.','Download test one- and two photon datasets.','Load Fiji/Miji into MATLAB path.','Download NWB (NeuroDataWithoutBorders)','Download EXTRACT'};
% Int vector: index of options.dependencyStr to run by default with no GUI
- options.depIdxArray = [1 2 3 5];
+ options.depIdxArray = [1 2 3 5 6];
% Binary: 1 = force update even if already downloaded. 0 = skip if already downloaded
options.forceUpdate = 0;
% get options
@@ -95,6 +96,16 @@ function loadDependencies(varargin)
optionsH.outputDir = optionsH.gitNameDisp;
optionsH.gitName = cellfun(@(x) [x '-master'],optionsH.gitNameDisp,'UniformOutput',false);
[success] = downloadGithubRepositories('options',optionsH);
+ case 'downloadEXTRACT'
+ optionsH.forceUpdate = forceUpdate;
+ optionsH.signalExtractionDir = options.externalProgramsDir;
+ optionsH.gitNameDisp = {'extract'};
+ optionsH.gitRepos = {'https://github.com/schnitzer-lab/EXTRACT-public'};
+ optionsH.gitRepos = cellfun(@(x) [x '/archive/master.zip'],optionsH.gitRepos,'UniformOutput',false);
+ optionsH.outputDir = optionsH.gitNameDisp;
+ % optionsH.gitName = cellfun(@(x) [x '-master'],optionsH.gitNameDisp,'UniformOutput',false);
+ optionsH.gitName = {'EXTRACT-public-master'};
+ [success] = downloadGithubRepositories('options',optionsH);
case 'downloadNeuroDataWithoutBorders'
optionsH.forceUpdate = forceUpdate;
optionsH.signalExtractionDir = options.externalProgramsDir;
diff --git a/+ciapkg/+io/loadSignalExtraction.m b/+ciapkg/+io/loadSignalExtraction.m
new file mode 100644
index 0000000..1e08480
--- /dev/null
+++ b/+ciapkg/+io/loadSignalExtraction.m
@@ -0,0 +1,288 @@
+function [inputImages,inputSignals,infoStruct,algorithmStr,inputSignals2] = loadSignalExtraction(inputFilePath,varargin)
+ % Loads CIAtah-style MAT or NWB files containing signal extraction results.
+ % Biafra Ahanonu
+ % started: 2021.02.03 [10:53:11]
+ % inputs
+ % inputFilePath - path to signal extraction output
+ % outputs
+ % inputImages - 3D or 4D matrix containing cells and their spatial information, format: [x y nCells].
+ % inputSignals - 2D matrix containing activity traces in [nCells nFrames] format.
+ % infoStruct - contains information about the file, e.g. the 'description' property that can contain information about the algorithm.
+ % algorithmStr - String of the algorithm name.
+ % inputSignals2 - same as inputSignals but for secondary traces an algorithm outputs.
+
+ % changelog
+ % 2021.03.10 [18:50:48] - Updated to add support for initial set of cell-extraction algorithms.
+ % TODO
+ %
+
+ % ========================
+ % Struct: name of structures for CIAtah-style outputs.
+ options.extractionMethodStructVarname = struct(...
+ 'PCAICA', 'pcaicaAnalysisOutput',...
+ 'EM', 'emAnalysisOutput',...
+ 'CELLMax','cellmaxAnalysisOutput',...
+ 'EXTRACT', 'extractAnalysisOutput',...
+ 'CNMF', 'cnmfAnalysisOutput',...
+ 'CNMFE', 'cnmfeAnalysisOutput',...
+ 'ROI', 'roiAnalysisOutput'...
+ );
+
+ options.extractionMethodStructSaveStr = struct(...
+ 'pcaicaAnalysis','PCAICA',...
+ 'emAnalysis','EM',...
+ 'cellmaxAnalysis','CELLMax',...
+ 'extractAnalysis','EXTRACT',...
+ 'cnmfAnalysis','CNMF',...
+ 'cnmfeAnalysis','CNMFE',...
+ 'roiAnalysis','ROI'...
+ );
+ % Name of H5 group for images and signal series in NWB files
+ options.nwbGroupImages = '/processing/ophys/ImageSegmentation/PlaneSegmentation';
+ options.nwbGroupSignalSeries = '/processing/ophys/Fluorescence/RoiResponseSeries';
+ % Int: indicates which trace output to use from 1 = primary trace, 2 = secondary trace, etc.
+ options.signalExtractionTraceOutputType = 1;
+ % get options
+ options = getOptions(options,varargin);
+ % display(options)
+ % unpack options into current workspace
+ % fn=fieldnames(options);
+ % for i=1:length(fn)
+ % eval([fn{i} '=options.' fn{i} ';']);
+ % end
+ % ========================
+
+ try
+ inputImages = [];
+ inputSignals = [];
+ inputSignals2 = [];
+ algorithmStr = '';
+ infoStruct = struct;
+
+ if iscell(inputFilePath)
+ inputFilePath = inputFilePath{1};
+ end
+ % Return if not a string, return as not appropriate input
+ if ~ischar(inputFilePath)
+ return;
+ end
+
+ [~,~,inputEXT] = fileparts(inputFilePath);
+ switch inputEXT
+ case '.nwb'
+ nwbOpts.groupImages = options.nwbGroupImages;
+ nwbOpts.groupSignalSeries = options.nwbGroupSignalSeries;
+ [inputImages,inputSignals,infoStruct,algorithmStr] = loadNeurodataWithoutBorders(inputFilePath,'options',nwbOpts);
+ return;
+ case '.mat'
+ % Do nothing, go to the next steps below.
+ otherwise
+ % Do nothing
+ end
+
+ % Get list of variables in the MAT file, use this to determine which algorithm to load.
+ variableInfo = who('-file', inputFilePath);
+
+ algList = fieldnames(options.extractionMethodStructVarname);
+ algorithm = '';
+ for i = 1:length(algList)
+ algName = algList{i};
+ algMatch = options.extractionMethodStructVarname.(algName);
+ matchAlg = any(strcmp(variableInfo,algMatch));
+ if matchAlg==1
+ algorithm = algName;
+ end
+ end
+ algorithmStr = algorithm;
+
+ switch algorithm
+ case 'ROI'
+ [inputImages,inputSignals,infoStruct] = subfxnROI(inputFilePath,options);
+ case 'PCAICA'
+ [inputImages,inputSignals,infoStruct] = subfxnPCAICA(inputFilePath,options);
+ case {'EM','CELLMax'}
+ [inputImages,inputSignals,infoStruct,inputSignals2] = subfxnCELLMax(inputFilePath,options);
+ case 'EXTRACT'
+ [inputImages,inputSignals,infoStruct] = subfxnEXTRACT(inputFilePath,options);
+ case 'CNMF'
+ [inputImages,inputSignals,infoStruct,inputSignals2] = subfxnCNMF(inputFilePath,options);
+ case 'CNMFE'
+ [inputImages,inputSignals,infoStruct,inputSignals2] = subfxnCNMFE(inputFilePath,options);
+ otherwise
+ disp('Input MAT file is not recognized or not in CIAtah-style format.')
+ end
+
+ catch err
+ disp(repmat('@',1,7))
+ disp(getReport(err,'extended','hyperlinks','on'));
+ disp(repmat('@',1,7))
+ end
+end
+function [inputImages,inputSignals,infoStruct] = subfxnPCAICA(inputFilePath,options)
+ infoStruct = struct;
+ load(inputFilePath);
+
+ if exist('pcaicaAnalysisOutput','var')
+ inputSignals = double(pcaicaAnalysisOutput.IcaTraces);
+ if strcmp(pcaicaAnalysisOutput.imageSaveDimOrder,'xyz')
+ % inputImages = permute(double(pcaicaAnalysisOutput.IcaFilters),[3 1 2])
+ inputImages = double(pcaicaAnalysisOutput.IcaFilters);
+ elseif strcmp(pcaicaAnalysisOutput.imageSaveDimOrder,'zxy')
+ inputImages = permute(double(pcaicaAnalysisOutput.IcaFilters),[2 3 1]);
+ % inputImages = pcaicaAnalysisOutput.IcaFilters;
+ else
+ % inputImages = permute(double(pcaicaAnalysisOutput.IcaFilters));
+ inputImages = pcaicaAnalysisOutput.IcaFilters;
+ end
+ end
+ if exist('IcaTraces','var')
+ inputSignals = IcaTraces;
+ end
+ if exist('IcaFilters','var')
+ % inputImages = IcaFilters;
+ inputImages = permute(IcaFilters,[2 3 1]);
+ end
+end
+function [inputImages,inputSignals,infoStruct,inputSignals2] = subfxnCELLMax(inputFilePath,options)
+ infoStruct = struct;
+ load(inputFilePath);
+
+ % if exist(options.structEMVarname,'var')
+ if exist('emAnalysisOutput','var')|exist('cellmaxAnalysisOutput','var')
+ if exist('cellmaxAnalysisOutput','var')
+ emAnalysisOutput = cellmaxAnalysisOutput;
+ end
+ inputImages = emAnalysisOutput.cellImages;
+
+ emOutputType = {'scaledProbability','dsScaledProbability','cellTraces','dsCellTraces'};
+ for emI = 1:length(emOutputType)
+ if isfield(emAnalysisOutput,emOutputType{emI})
+ disp(['Using ' emOutputType{emI} '...'])
+ inputSignals = double(emAnalysisOutput.(emOutputType{emI}));
+ break;
+ end
+ end
+
+ if 0
+ if isfield(emAnalysisOutput,'scaledProbability')
+ disp('Using scaledProbability...')
+ inputSignals = double(emAnalysisOutput.scaledProbability);
+ elseif isfield(emAnalysisOutput,'dsScaledProbability')
+ disp('Using dsScaledProbability...')
+ inputSignals = double(emAnalysisOutput.dsScaledProbability);
+ elseif isfield(emAnalysisOutput,'cellTraces')
+ disp('Using cellTraces...')
+ inputSignals = double(emAnalysisOutput.cellTraces);
+ elseif isfield(emAnalysisOutput,'dsCellTraces')
+ disp('Using dsCellTraces...')
+ if length(emAnalysisOutput.dsCellTraces)==1
+ inputSignals = emAnalysisOutput.cellTraces;
+ else
+ inputSignals = emAnalysisOutput.dsCellTraces;
+ end
+ else
+ inputSignals = emAnalysisOutput.cellTraces;
+ end
+ end
+
+ inputSignals2 = double(emAnalysisOutput.cellTraces);
+
+ switch options.signalExtractionTraceOutputType
+ case 1
+ %
+ case 2
+ inputSignals2Tmp = inputSignals2;
+ inputSignals2 = inputSignals;
+ inputSignals = inputSignals2Tmp;
+ otherwise
+ %
+ end
+
+ % Convert to dF/F values
+ inputSignals = normalizeSignalExtractionActivityTraces(inputSignals,inputImages);
+ inputSignals2 = normalizeSignalExtractionActivityTraces(inputSignals2,inputImages);
+
+ end
+end
+function [inputImages,inputSignals,infoStruct] = subfxnEXTRACT(inputFilePath,options)
+ infoStruct = struct;
+ load(inputFilePath);
+
+ % if exist(options.structEXTRACTVarname,'var')
+ if exist('extractAnalysisOutput','var')
+ % inputImages = double(permute(extractAnalysisOutput.filters,[3 1 2]));
+ inputImages = double(extractAnalysisOutput.filters);
+ % inputSignals = double(permute(extractAnalysisOutput.traces, [2 1]));
+ inputSignals = double(extractAnalysisOutput.traces);
+ end
+end
+function [inputImages,inputSignals,infoStruct,inputSignals2] = subfxnCNMF(inputFilePath,options)
+ infoStruct = struct;
+ load(inputFilePath);
+
+ % if exist(options.structCNMRVarname,'var')
+ if exist('cnmfAnalysisOutput','var')
+ % inputImages = double(permute(cnmfAnalysisOutput.extractedImages,[3 1 2]));
+ inputImages = double(cnmfAnalysisOutput.extractedImages);
+ % inputSignals = double(permute(extractAnalysisOutput.traces, [2 1]));
+
+ switch options.signalExtractionTraceOutputType
+ case 1
+ inputSignals = double(cnmfAnalysisOutput.extractedSignals);
+ inputSignals2 = double(cnmfAnalysisOutput.extractedSignalsEst);
+ case 2
+ inputSignals = double(cnmfAnalysisOutput.extractedSignalsEst);
+ inputSignals2 = double(cnmfAnalysisOutput.extractedSignals);
+ otherwise
+ inputSignals = double(cnmfAnalysisOutput.extractedSignalsEst);
+ inputSignals2 = double(cnmfAnalysisOutput.extractedSignals);
+ end
+
+ % Convert units to relative dF/F for later analysis
+ inputSignals = normalizeSignalExtractionActivityTraces(inputSignals,inputImages);
+ inputSignals2 = normalizeSignalExtractionActivityTraces(inputSignals2,inputImages);
+ end
+end
+function [inputImages,inputSignals,infoStruct,inputSignals2] = subfxnCNMFE(inputFilePath,options)
+ infoStruct = struct;
+ load(inputFilePath);
+
+ if exist('cnmfeAnalysisOutput','var')
+ inputImages = double(cnmfeAnalysisOutput.extractedImages);
+
+ switch options.signalExtractionTraceOutputType
+ case 1
+ inputSignals = double(cnmfeAnalysisOutput.extractedSignals);
+ inputSignals2 = double(cnmfeAnalysisOutput.extractedSignalsEst);
+ case 2
+ inputSignals = double(cnmfeAnalysisOutput.extractedSignalsEst);
+ inputSignals2 = double(cnmfeAnalysisOutput.extractedSignals);
+ otherwise
+ inputSignals = double(cnmfeAnalysisOutput.extractedSignalsEst);
+ inputSignals2 = double(cnmfeAnalysisOutput.extractedSignals);
+ end
+
+ % Convert units to relative dF/F for later analysis
+ inputSignals = normalizeSignalExtractionActivityTraces(inputSignals,inputImages);
+ inputSignals2 = normalizeSignalExtractionActivityTraces(inputSignals2,inputImages);
+ end
+end
+function [inputImages,inputSignals,infoStruct] = subfxnROI(inputFilePath,options)
+ infoStruct = struct;
+ load(inputFilePath);
+
+ if exist('roiAnalysisOutput','var')
+ % inputImages = double(permute(extractAnalysisOutput.filters,[3 1 2]));
+ inputImages = double(roiAnalysisOutput.filters);
+ % inputSignals = double(permute(extractAnalysisOutput.traces, [2 1]));
+ if isfield(roiAnalysisOutput,'tracesDemix')
+ display('Using demixed ROI traces!')
+ inputSignals = double(roiAnalysisOutput.tracesDemix);
+ else
+ inputSignals = double(roiAnalysisOutput.traces);
+ end
+ end
+ if exist('ROItraces','var')
+ inputSignals = ROItraces;
+ end
+end
\ No newline at end of file
diff --git a/+ciapkg/+io/saveSignalExtraction.m b/+ciapkg/+io/saveSignalExtraction.m
new file mode 100644
index 0000000..0c1ec8e
--- /dev/null
+++ b/+ciapkg/+io/saveSignalExtraction.m
@@ -0,0 +1,263 @@
+function [success] = saveSignalExtraction(inputImages,inputSignals,signalExtractionMethod,outputFilePath,varargin)
+ % Convert signal extraction outputs to CIAtah-style MAT files.
+ % Biafra Ahanonu
+ % started: 2021.02.02 [14:24:35]
+ % inputs
+ % Matrix: inputImages - [x y z] matrix
+ % Matrix: inputSignals - {1 N} cell with N = number of different signal traces for that algorithm. Make sure each signal trace matrix is in form of [nSignals nFrames].
+ % Str: signalExtractionMethod - Name of the signal-extraction method used. Options: CELLMax, PCAICA, CELLMax, EXTRACT, CNMF, CNMFE, ROI.
+ % Str: outputFilePath - file path to save MAT file to. This function will automatically add the appropriate suffix and extension.
+ % outputs
+ % success - alerts user of saving of files was successful.
+
+ % changelog
+ % 2021.03.20 [20:35:50] - Adding support for all existing methods.
+ % TODO
+ %
+
+ % ========================
+ % Str: Name of the movie that signal extraction was performed on.
+ options.movieList = '';
+ % Matrix: 2D matrix of [nCells nFrames], secondary input activity trace if needed.
+ options.inputSignals2 = [];
+ % Struct: name of structures for CIAtah-style outputs.
+ options.extractionMethodStructSaveStr = struct(...
+ 'PCAICA', '_pcaicaAnalysis.mat',...
+ 'EM', '_emAnalysis.mat',...
+ 'CELLMax', '_cellmaxAnalysis.mat',...
+ 'EXTRACT', '_extractAnalysis.mat',...
+ 'CNMF', '_cnmfAnalysis.mat',...
+ 'CNMFE', '_cnmfeAnalysis.mat',...
+ 'ROI', '_roiAnalysis.mat'...
+ );
+ % Struct: name of structures for CIAtah-style outputs.
+ options.extractionMethodStructVarname = struct(...
+ 'PCAICA', 'pcaicaAnalysisOutput',...
+ 'EM', 'emAnalysisOutput',...
+ 'CELLMax', 'cellmaxAnalysisOutput',...
+ 'EXTRACT', 'extractAnalysisOutput',...
+ 'CNMF', 'cnmfAnalysisOutput',...
+ 'CNMFE', 'cnmfeAnalysisOutput',...
+ 'ROI', 'roiAnalysisOutput'...
+ );
+ % INTERNAL
+ % Str: name of algorithm being analyzed
+ options.signalExtractionMethod = '';
+
+ % get options
+ options = getOptions(options,varargin);
+ % display(options)
+ % unpack options into current workspace
+ % fn=fieldnames(options);
+ % for i=1:length(fn)
+ % eval([fn{i} '=options.' fn{i} ';']);
+ % end
+ % ========================
+
+ try
+ success = 0;
+ if ~strcmp(signalExtractionMethod,'ROI')
+ options.signalExtractionMethod = signalExtractionMethod;
+ end
+
+ [filePath,fileName,ext] = fileparts(outputFilePath);
+
+ try
+ % Append the expected CIAtah-style suffix and MAT file extension.
+ outputFilePath = [filePath filesep fileName options.extractionMethodStructSaveStr.(signalExtractionMethod)];
+ catch err
+ disp(repmat('@',1,7))
+ disp(getReport(err,'extended','hyperlinks','on'));
+ disp(repmat('@',1,7))
+ end
+
+ switch signalExtractionMethod
+ case 'ROI'
+ subfxnROI(inputImages,inputSignals,signalExtractionMethod,outputFilePath,options);
+ case 'PCAICA'
+ subfxnPCAICA(inputImages,inputSignals,signalExtractionMethod,outputFilePath,options);
+ case {'EM','CELLMax'}
+ subfxnCELLMax(inputImages,inputSignals,signalExtractionMethod,outputFilePath,options);
+ case 'EXTRACT'
+ subfxnEXTRACT(inputImages,inputSignals,signalExtractionMethod,outputFilePath,options);
+ case 'CNMF'
+ subfxnCNMF(inputImages,inputSignals,signalExtractionMethod,outputFilePath,options);
+ case 'CNMFE'
+ subfxnCNMFE(inputImages,inputSignals,signalExtractionMethod,outputFilePath,options);
+ otherwise
+ disp('Input MAT file is not recognized or not in CIAtah-style format.')
+ success = 0;
+ return;
+ end
+ success = 1;
+ catch err
+ success = 0;
+ disp(repmat('@',1,7))
+ disp(getReport(err,'extended','hyperlinks','on'));
+ disp(repmat('@',1,7))
+ end
+end
+function subfxnSaveOutput(inputOutputStruct,additonalStruct,outputFilePath,options)
+ % Save using that algorithms structure name
+ structSaveName = options.extractionMethodStructVarname.(options.signalExtractionMethod);
+ tmpStruct.(structSaveName) = inputOutputStruct;
+ if ~isempty(additonalStruct)
+ tmpStruct.(additonalStruct{1}) = additonalStruct{2};
+ end
+ % =======
+ % save output components
+ saveID = {'1'};
+ thisDirSaveStr = outputFilePath;
+ for i=1:length(saveID)
+ savestring = [thisDirSaveStr saveID{i}];
+ display(['Saving: ' outputFilePath])
+ save(outputFilePath,'-struct', 'tmpStruct','-v7.3');
+ end
+end
+function subfxnROI(inputImages,inputSignals,signalExtractionMethod,outputFilePath,options)
+ roiAnalysisOutput.filters = inputImages;
+ roiAnalysisOutput.traces = inputSignals;
+ roiAnalysisOutput.signalExtractionMethod = options.signalExtractionMethod;
+
+ options.signalExtractionMethod = signalExtractionMethod;
+
+ subfxnSaveOutput(roiAnalysisOutput,[],outputFilePath,options);
+end
+function subfxnPCAICA(inputImages,inputSignals,signalExtractionMethod,outputFilePath,options)
+ %
+
+ imageSaveDimOrder = 'xyz';
+ traceSaveDimOrder = '[nComponents frames]';
+
+ pcaicaAnalysisOutput.IcaFilters = inputImages;
+ pcaicaAnalysisOutput.IcaTraces = inputSignals;
+
+ pcaicaAnalysisOutput.imageSaveDimOrder = imageSaveDimOrder;
+ pcaicaAnalysisOutput.traceSaveDimOrder = traceSaveDimOrder;
+ pcaicaAnalysisOutput.nPCs = NaN;
+ pcaicaAnalysisOutput.nICs = size(inputImages,3);
+ pcaicaAnalysisOutput.time.startTime = NaN; %startTime;
+ pcaicaAnalysisOutput.time.endTime = NaN; %toc(startTime);
+ pcaicaAnalysisOutput.time.dateTime = NaN; %datestr(now,'yyyymmdd_HHMM','local');
+ pcaicaAnalysisOutput.movieList = options.movieList;
+ pcaicaAnalysisOutput.IcaInfo = []; % IcaInfo;
+
+ subfxnSaveOutput(pcaicaAnalysisOutput,[],outputFilePath,options);
+end
+function subfxnCELLMax(inputImages,inputSignals,signalExtractionMethod,outputFilePath,options)
+ %
+ currentDateTimeStr = datestr(now,'yyyymmdd_HHMM','local');
+
+ cellmaxAnalysisOutput.movieFilename = options.movieList;
+
+ cellmaxAnalysisOutput.cellImages = inputImages;
+ cellmaxAnalysisOutput.scaledProbability = inputSignals;
+ cellmaxAnalysisOutput.cellTraces = options.inputSignals2;
+
+ cellmaxAnalysisOutput.scaledProbabilityDff = [];
+ cellmaxAnalysisOutput.cellTracesDff = [];
+
+ cellmaxAnalysisOutput.versionCellmax = '';
+ cellmaxAnalysisOutput.runCompletedNoErrors = 1;
+ cellmaxAnalysisOutput.movieDims = NaN;
+ cellmaxAnalysisOutput.centroids = [];
+ cellmaxAnalysisOutput.CELLMaxoptions = struct;
+ cellmaxAnalysisOutput.filtTraces = [];
+ cellmaxAnalysisOutput.eventTimes = {};
+ cellmaxAnalysisOutput.eventOptions = struct;
+ cellmaxAnalysisOutput.runtime = NaN;
+ cellmaxAnalysisOutput.runtimeWithIO = NaN;
+ cellmaxAnalysisOutput.options = struct;
+
+ % Secondary options structure
+ emOptions.CELLMaxoptions.numSignalsDetected = size(inputSignals,1);
+
+ emOptions.CELLMaxoptions.sqSizeX = [];
+ emOptions.CELLMaxoptions.sqSizeY = [];
+ emOptions.versionCellmax = NaN;
+ emOptions.time.startTime = NaN;
+ emOptions.time.endTime = NaN;
+ emOptions.time.cellmaxRuntime = NaN;
+ emOptions.time.cellmaxRuntime = NaN;
+
+ subfxnSaveOutput(cellmaxAnalysisOutput,{'emOptions',emOptions},outputFilePath,options);
+end
+function subfxnEXTRACT(inputImages,inputSignals,signalExtractionMethod,outputFilePath,options)
+
+ extractAnalysisOutput.filters = inputImages;
+ % MAKE SURE TRANSPOSED
+ disp('Note: make sure EXTRACT traces are transposed to [# of cells x # of frames] instead of EXTRACT default format [# of frames x # of cells]')
+ extractAnalysisOutput.traces = inputSignals;
+
+ extractAnalysisOutput.file = options.movieList;
+
+ extractAnalysisOutput.info = struct;
+ extractAnalysisOutput.config = struct;
+ extractAnalysisOutput.info = struct;
+
+ % Remove the large summary field since takes up unnecessary space
+ extractAnalysisOutput.info.summary = [];
+ extractAnalysisOutput.userInputConfig = struct;
+
+ % for backwards compatibility
+ extractAnalysisOutput.opts = struct;
+ extractAnalysisOutput.time.startTime = NaN;
+ extractAnalysisOutput.time.endTime = NaN;
+ extractAnalysisOutput.time.totalTime = NaN;
+
+ subfxnSaveOutput(extractAnalysisOutput,{},outputFilePath,options);
+end
+function subfxnCNMF(inputImages,inputSignals,signalExtractionMethod,outputFilePath,options)
+ cnmfAnalysisOutput.extractedImages = inputImages;
+ % correct for df/f output problems
+ cnmfAnalysisOutput.extractedSignals = inputSignals;
+ cnmfAnalysisOutput.extractedSignalsEst = options.inputSignals2;
+
+ % add parameters and extractions to output structure
+ cnmfAnalysisOutput.params.K = NaN;
+ cnmfAnalysisOutput.params.tau = NaN;
+ cnmfAnalysisOutput.params.p = NaN;
+ cnmfAnalysisOutput.datasetComponentProperties_P = NaN;
+ cnmfAnalysisOutput.movieList = options.movieList;
+
+ cnmfAnalysisOutput.extractedSignalsBackground = NaN;
+ cnmfAnalysisOutput.extractedSignalsType = 'dfof';
+ cnmfAnalysisOutput.extractedSignalsEstType = 'model';
+ cnmfAnalysisOutput.extractedPeaks = NaN;
+ cnmfAnalysisOutput.extractedPeaksEst = NaN;
+ cnmfAnalysisOutput.cnmfOptions = struct;
+ cnmfAnalysisOutput.time.datetime = '';
+ cnmfAnalysisOutput.time.runtimeWithMovie = NaN;
+ cnmfAnalysisOutput.time.runtimeSansMovie = NaN;
+
+ cnmfAnalysisOutput.time.startTime = NaN;
+ cnmfAnalysisOutput.time.endTime = NaN;
+ cnmfAnalysisOutput.time.totalTime = NaN;
+
+ cnmfAnalysisOutput.versionOutput = 'current';
+
+ subfxnSaveOutput(cnmfAnalysisOutput,{},outputFilePath,options);
+end
+function subfxnCNMFE(inputImages,inputSignals,signalExtractionMethod,outputFilePath,options)
+ cnmfeAnalysisOutput.extractedImages = inputImages;
+ cnmfeAnalysisOutput.extractedSignals = inputSignals;
+ cnmfeAnalysisOutput.extractedSignalsEst = options.inputSignals2;
+
+ cnmfeAnalysisOutput.success = 1;
+ cnmfeAnalysisOutput.params = NaN;
+ cnmfeAnalysisOutput.movieList = options.movieList;
+ % cnmfeAnalysisOutput.extractedImages = reshape(full(results.A),[size(results.P.sn) size(results.C,1)]);
+
+ cnmfeAnalysisOutput.extractedSignalsType = 'model';
+ cnmfeAnalysisOutput.extractedSignalsEstType = 'dfof';
+ cnmfeAnalysisOutput.extractedPeaks = [];
+ cnmfeAnalysisOutput.Cn = [];
+ cnmfeAnalysisOutput.P = [];
+
+ cnmfeAnalysisOutput.time.startTime = NaN;
+ cnmfeAnalysisOutput.time.endTime = NaN;
+ cnmfeAnalysisOutput.time.totalTime = NaN;
+ cnmfeAnalysisOutput.obj.cnmfeOptions = struct;
+
+ subfxnSaveOutput(cnmfeAnalysisOutput,{},outputFilePath,options);
+end
\ No newline at end of file
diff --git a/+ciapkg/+io/updatePkg.m b/+ciapkg/+io/updatePkg.m
index 1a9c6fb..dc4adad 100644
--- a/+ciapkg/+io/updatePkg.m
+++ b/+ciapkg/+io/updatePkg.m
@@ -8,7 +8,7 @@
%
% changelog
- %
+ % 2021.03.09 [21:34:52] - Option on whether to update package (only 0 for now). Also alert user if behind a version.
% TODO
%
@@ -24,6 +24,8 @@
options.versionURL = 'https://api.github.com/repos/bahanonu/calciumImagingAnalysis/contents/ciapkg/VERSION';
% Binary: 1 = pop-up GUI enabled
options.showGui = 1;
+ % Binary: 1 = update code, 0 = only check for update and notify user behind a version.
+ options.updatePackage = 0;
% get options
options = getOptions(options,varargin);
% display(options)
@@ -49,10 +51,28 @@
if verCompare==1
verInfoStr = 'Running most up-to-date version!';
+ verInfoStr2 = 'Running most up-to-date version!';
elseif verCompare<1
verInfoStr = 'Running behind! Initiating update [IGNORE for now].';
+ verInfoStr2 = 'Running a version behind, consider updating.';
elseif verCompare>1
verInfoStr = 'I do not know how, but you are running a version ahead! [Dev build]';
+ verInfoStr2 = 'I do not know how, but you are running a version ahead! [Dev build]';
+ end
+
+ if options.updatePackage==0
+ if iscell(onlineVersion)
+ onlineVersionTmp = onlineVersion{1};
+ else
+ onlineVersionTmp = onlineVersion;
+ end
+ verInfoStr = sprintf('%s\n Local version: %s.\n Online version %s.\n',verInfoStr2,currentVersion,onlineVersionTmp);
+ warning(verInfoStr)
+ if verCompare<1
+ msgbox(verInfoStr,'Note on CIAtah version.')
+ end
+ success = 1;
+ return
end
disp(verInfoStr)
@@ -81,6 +101,12 @@
end
function [verCompare,compareVector] = subfxn_verCompare(verId1,verId2)
fprintf('Comparing\nLocal: %s.\nOnline: %s.\n',verId1,verId2);
+ if iscell(verId1)
+ verId1 = verId1{1};
+ end
+ if iscell(verId2)
+ verId2 = verId2{1};
+ end
% Remove version tag
verId1 = strrep(verId1,'v','');
verId2 = strrep(verId2,'v','');
@@ -89,7 +115,7 @@
verId1 = cellfun(@(x) str2num(x),strsplit(verId1,'.'));
verId2 = cellfun(@(x) str2num(x),strsplit(verId2,'.'));
compareVector = zeros([1 max(length(verId1),length(verId2))]);
- nLevels = length(compareVector)
+ nLevels = length(compareVector);
% Start at the highest level and only continue comparing while version 2 is greater than or equal to version 1.
lockOut = 0; % 1 = higher level is an old version, so only calculate version difference
@@ -110,6 +136,6 @@
if sum(compareVector)>0
verCompare = 2;
end
- disp(compareVector)
+ % disp(compareVector)
end
end
\ No newline at end of file
diff --git a/+ciapkg/pkgName.m b/+ciapkg/pkgName.m
new file mode 100644
index 0000000..d344d1a
--- /dev/null
+++ b/+ciapkg/pkgName.m
@@ -0,0 +1,36 @@
+function [pkgNameStr] = pkgName(varargin)
+ % Get name for the package (CIAtah by default).
+ % Biafra Ahanonu
+ % started: updated: 2021.03.16 [14:39:41]
+ % inputs
+ %
+ % outputs
+ %
+
+ % changelog
+ %
+ % TODO
+ %
+
+ %========================
+ % DESCRIPTION
+ % options.exampleOption = '';
+ % get options
+ % options = getOptions(options,varargin);
+ % display(options)
+ % unpack options into current workspace
+ % fn=fieldnames(options);
+ % for i=1:length(fn)
+ % eval([fn{i} '=options.' fn{i} ';']);
+ % end
+ %========================
+
+ try
+ pkgNameStr = 'CIAtah';
+ catch err
+ pkgNameStr = 'CIAtah';
+ disp(repmat('@',1,7))
+ disp(getReport(err,'extended','hyperlinks','on'));
+ disp(repmat('@',1,7))
+ end
+end
\ No newline at end of file
diff --git a/+ciapkg/versionOnline.m b/+ciapkg/versionOnline.m
index 3ebdd78..af0cd59 100644
--- a/+ciapkg/versionOnline.m
+++ b/+ciapkg/versionOnline.m
@@ -35,9 +35,11 @@
% Get version information online
% Get information about specific version file online using GitHub API
[versionInfo, status] = urlread(options.versionURL,'Timeout',options.timeOutSec);
+ % [versionInfo, status] = webread(options.versionURL,'Timeout',options.timeOutSec);
if status==1
versionInfo = jsondecode(versionInfo);
[onlineVersion, status] = urlread(versionInfo.download_url,'Timeout',options.timeOutSec);
+ % [onlineVersion, status] = webread(versionInfo.download_url,'Timeout',options.timeOutSec);
if status==0
disp('Could not dowload CIAPKG version information.')
return;
diff --git a/@calciumImagingAnalysis/calciumImagingAnalysis.m b/@calciumImagingAnalysis/calciumImagingAnalysis.m
index d7b3e4d..c47889c 100644
--- a/@calciumImagingAnalysis/calciumImagingAnalysis.m
+++ b/@calciumImagingAnalysis/calciumImagingAnalysis.m
@@ -174,7 +174,7 @@
extractionMethodSaveStr = struct(...
'PCAICA', '_pcaicaAnalysis',...
'EM', '_emAnalysis',...
- 'CELLMax','_cellmaxAnalysis',...
+ 'CELLMax', '_cellmaxAnalysis',...
'EXTRACT', '_extractAnalysis',...
'CNMF', '_cnmfAnalysis',...
'CNMFE', '_cnmfeAnalysis',...
@@ -183,6 +183,7 @@
extractionMethodStructSaveStr = struct(...
'PCAICA', '_pcaicaAnalysis.mat',...
'EM', '_emAnalysis.mat',...
+ 'CELLMax', '_cellmaxAnalysis.mat',...
'EXTRACT', '_extractAnalysis.mat',...
'CNMF', '_cnmfAnalysis.mat',...
'CNMFE', '_cnmfeAnalysis.mat',...
@@ -191,6 +192,7 @@
extractionMethodSortedSaveStr = struct(...
'PCAICA', '_ICdecisions.mat',...
'EM', '_emAnalysisSorted.mat',...
+ 'CELLMax', '_cellmaxAnalysisSorted.mat',...
'EXTRACT', '_extractAnalysisSorted.mat',...
'CNMF', '_cnmfAnalysisSorted.mat',...
'CNMFE', '_cnmfeAnalysisSorted.mat',...
@@ -199,6 +201,7 @@
extractionMethodClassifierSaveStr = struct(...
'PCAICA', '_ICclassifierDecisions.mat',...
'EM', '_emAnalysisClassifierDecisions.mat',...
+ 'CELLMax', '_cellmaxAnalysisClassifierDecisions.mat',...
'EXTRACT', '_extractAnalysisClassifierDecisions.mat',...
'CNMF', '_cnmfAnalysisClassifierDecisions.mat',...
'CNMFE', '_cnmfeAnalysisClassifierDecisions.mat',...
@@ -207,6 +210,7 @@
extractionMethodValidVarname = struct(...
'PCAICA', 'valid',...
'EM', 'validCellMax',...
+ 'CELLMax', 'validCellMax',...
'EXTRACT', 'validEXTRACT',...
'CNMF', 'validCNMF',...
'CNMFE', 'validCNMFE',...
@@ -215,6 +219,7 @@
extractionMethodStructVarname = struct(...
'PCAICA', 'pcaicaAnalysisOutput',...
'EM', 'emAnalysisOutput',...
+ 'CELLMax', 'cellmaxAnalysisOutput',...
'EXTRACT', 'extractAnalysisOutput',...
'CNMF', 'cnmfAnalysisOutput',...
'CNMFE', 'cnmfeAnalysisOutput',...
@@ -709,7 +714,7 @@
% these are in separate M-files
% calciumImagingAnalysis main class methods
- [idNumIdxArray, validFoldersIdx, ok] = calciumImagingAnalysisMainGui(obj,fxnsToRun,inputTxt,currentIdx)
+ [idNumIdxArray, validFoldersIdx, ok] = ciatahMainGui(obj,fxnsToRun,inputTxt,currentIdx)
[idNumIdxArray, ok] = pipelineListBox(obj,fxnsToRun,inputTxt,currentIdx)
obj = computeCrossDayDistancesAlignment(obj)
obj = computeCellDistances(obj)
diff --git a/@calciumImagingAnalysis/calciumImagingAnalysisMainGui.m b/@calciumImagingAnalysis/ciatahMainGui.m
similarity index 92%
rename from @calciumImagingAnalysis/calciumImagingAnalysisMainGui.m
rename to @calciumImagingAnalysis/ciatahMainGui.m
index 855f1e6..cb0bc94 100644
--- a/@calciumImagingAnalysis/calciumImagingAnalysisMainGui.m
+++ b/@calciumImagingAnalysis/ciatahMainGui.m
@@ -1,4 +1,4 @@
-function [idNumIdxArray, validFoldersIdx, ok] = calciumImagingAnalysisMainGui(obj,fxnsToRun,inputTxt,currentIdx)
+function [idNumIdxArray, validFoldersIdx, ok] = ciatahMainGui(obj,fxnsToRun,inputTxt,currentIdx)
% Main GUI for calciumImagingAnalysis startup
% Biafra Ahanonu
% started: 2020.03.23 [22:36:36] - branch from calciumImagingAnalysis 2020.05.07 [15:47:29]
@@ -44,8 +44,8 @@
hFig = figure;
hListboxS = struct;
- set(hFig,'Name','CIAtah: start-up GUI','NumberTitle','off')
- uicontrol('Style','text','String',['CIAtah'],'Units','normalized','Position',[1 97.5 20 2.5]/100,'BackgroundColor','white','HorizontalAlignment','Left','ForegroundColor','black','FontWeight','bold','FontAngle','italic');
+ set(hFig,'Name',[ciapkg.pkgName ': start-up GUI'],'NumberTitle','off')
+ uicontrol('Style','text','String',[ciapkg.pkgName],'Units','normalized','Position',[1 97.5 20 2.5]/100,'BackgroundColor','white','HorizontalAlignment','Left','ForegroundColor','black','FontWeight','bold','FontAngle','italic');
uicontrol('Style','text','String',[inputTxt 10 'Press TAB to select next section, ENTER to continue, and ESC to exit.'],'Units','normalized','Position',[10 92 90 8]/100,'BackgroundColor','white','HorizontalAlignment','Left','ForegroundColor','black');
% set(hFig,'Color',[0,0,0]);
@@ -74,14 +74,14 @@
selBoxInfo.methods.string = fxnsToRun;
selBoxInfo.cellExtract.string = usrIdxChoiceDisplay;
- selBoxInfo.cellExtractFiletype.string = {'CIAtah format','NeuroDataWithoutBorders (NWB) format'};
+ selBoxInfo.cellExtractFiletype.string = {[ciapkg.pkgName ' format'],'NeuroDataWithoutBorders (NWB) format'};
selBoxInfo.folderFilt.string = useAltValid;
selBoxInfo.subject.string = subjectStrUnique;
selBoxInfo.assay.string = assayStrUnique;
selBoxInfo.folders.string = selectList;
selBoxInfo.guiEnabled.string = {'GUI in methods enabled','GUI in methods disabled'};
- selBoxInfo.methods.title = 'Select a CIAtah method:';
+ selBoxInfo.methods.title = ['Select a ' ciapkg.pkgName ' method:'];
selBoxInfo.cellExtract.title = 'Cell-extraction method:';
selBoxInfo.cellExtractFiletype.title = 'Cell-extraction file format:';
selBoxInfo.folderFilt.title = 'Folder select filters:';
@@ -335,12 +335,16 @@ function onKeyPressRelease(src, evnt, pressRelease,hFig)
% If NWB chosen as file-type, verify user has correct NWB inputs
if any(strcmp('cellExtractFiletypeBox',get(src,'Tag')))&nwbSetMovieInfoSwitch==0
- get(hListboxS.cellExtractFiletype,'Value')
+ disp(['cellExtractFiletype: ' num2str(get(hListboxS.cellExtractFiletype,'Value'))])
if get(hListboxS.cellExtractFiletype,'Value')==2
nwbSetMovieInfoSwitch = 1;
checkEnabled = 0;
- uiwait(msgbox(['Please enter information for NWB cell-extraction and imaging movie regular expressions (for locating files) and whether cell-extraction files are located in a sub-folder within each folder.' 10 10 'Press OK/enter to continue.'],'Note to user','modal'));
+ dispStr = 'Please enter information for NWB cell-extraction and imaging movie regular expressions (for locating files) and whether cell-extraction files are located in a sub-folder within each folder.';
+ disp(dispStr)
+ disp('If Matlab UI is non-responsive, make sure you have checked for and closed pop-up dialog boxes.')
+ uiwait(msgbox([dispStr 10 10 'Press OK/enter to continue.'],'Note to user','modal'));
obj.setMovieInfo;
+ disp(['NWB information set! Remember to select the ' ciapkg.pkgName ' GUI then press Enter to start a module.'])
pause(0.1);
checkEnabled = 1;
end
diff --git a/@calciumImagingAnalysis/computeManualSortSignals.m b/@calciumImagingAnalysis/computeManualSortSignals.m
index 8bdcaae..732d51c 100644
--- a/@calciumImagingAnalysis/computeManualSortSignals.m
+++ b/@calciumImagingAnalysis/computeManualSortSignals.m
@@ -12,6 +12,7 @@
% 2019.05.15 [13:13:10] Added support for reading movie from disk to allow sorting of large imaging movies
% 2019.09.09 [18:14:48] - Update to handling uigetfile output.
% 2021.01.21 [10:37:07] - Updated to support HDF5 and regexp for movie manual names along with misc. other changes.
+ % 2021.03.17 [16:34:59] - If user hasn't called modelVarsFromFiles, computeManualSortSignals called the function. However, this lead to a mismatch between computeManualSortSignals fileNum and obj.fileNum, leading to mismatch between xcoords, etc. and input signals/images.
% TODO
% ADD PERSONS NAME TO THE FILE
@@ -38,7 +39,7 @@
obj.fileNum = fileNum;
% fileNum = obj.fileNum;
display(repmat('#',1,21))
- display([num2str(thisFileNumIdx) '/' num2str(nFilesToAnalyze) ' (' num2str(fileNum) '/' num2str(nFiles) '): ' obj.fileIDNameArray{obj.fileNum}]);
+ display([num2str(thisFileNumIdx) '/' num2str(nFilesToAnalyze) ' (' num2str(obj.fileNum) '/' num2str(nFiles) '): ' obj.fileIDNameArray{obj.fileNum}]);
% =======
% path to current folder
currentFolderPath = obj.inputFolders{obj.fileNum};
@@ -98,7 +99,9 @@
skipReload = 1;
catch
obj.guiEnabled = 0;
+ % originalFileNum =
obj.modelVarsFromFiles();
+ obj.fileNum = fileNum;
obj.guiEnabled = 1;
skipReload = 0;
end
@@ -290,7 +293,7 @@
% ioptions.inputStr = subjAssayIDStr;
ioptions.valid = valid;
ioptions.sessionID = [folderBaseSaveStr '_' num2str(java.lang.System.currentTimeMillis)];
- ioptions.inputStr = [num2str(thisFileNumIdx) '/' num2str(nFilesToAnalyze) ' (' num2str(fileNum) '/' num2str(nFiles) '): ' obj.folderBaseSaveStr{obj.fileNum}];
+ ioptions.inputStr = [num2str(thisFileNumIdx) '/' num2str(nFilesToAnalyze) ' (' num2str(obj.fileNum) '/' num2str(nFiles) '): ' obj.folderBaseSaveStr{obj.fileNum}];
ioptions.showROITrace = usrIdxChoiceROI;
ioptions.cropSizeLength = usrIdxCropSizeLength;
ioptions.cropSize = usrIdxCropSizeLength;
@@ -298,8 +301,8 @@
ioptions.thresholdOutline = userIdxImageThresholdOutline;
ioptions.colormap = obj.colormap;
- ioptions.coord.xCoords = max(1,round(obj.objLocations{fileNum}.(obj.signalExtractionMethod)(:,1)));
- ioptions.coord.yCoords = max(1,round(obj.objLocations{fileNum}.(obj.signalExtractionMethod)(:,2)));
+ ioptions.coord.xCoords = max(1,round(obj.objLocations{obj.fileNum}.(obj.signalExtractionMethod)(:,1)));
+ ioptions.coord.yCoords = max(1,round(obj.objLocations{obj.fileNum}.(obj.signalExtractionMethod)(:,2)));
ioptions.inputSignalsSecond = rawSignals2;
diff --git a/@calciumImagingAnalysis/loadDependencies.m b/@calciumImagingAnalysis/loadDependencies.m
index af09f91..1505f48 100644
--- a/@calciumImagingAnalysis/loadDependencies.m
+++ b/@calciumImagingAnalysis/loadDependencies.m
@@ -12,17 +12,18 @@
% 2020.06.28 [14:25:04] - Added ability for users to force update.
% 2021.01.22 [13:42:36] - NWB from specific release to reduce compatibility errors.
% 2021.02.01 [15:10:41] - Calls non-class function for use in more functions without needing to load CIAtah class.
+ % 2021.03.20 [18:12:20] - Added EXTRACT support to list of functions to download.
% TODO
% Verify all dependencies download and if not ask user to download again.
%========================
% DESCRIPTION
options.guiEnabled = 1;
- options.dependencyStr = {'downloadMiji','downloadCnmfGithubRepositories','example_downloadTestData','loadMiji','downloadNeuroDataWithoutBorders'};
+ options.dependencyStr = {'downloadMiji','downloadCnmfGithubRepositories','example_downloadTestData','loadMiji','downloadNeuroDataWithoutBorders','downloadEXTRACT'};
- options.dispStr = {'Download Fiji (to run Miji)','Download CNMF, CNMF-E, and CVX code.','Download test one- and two photon datasets.','Load Fiji/Miji into MATLAB path.','Download NWB (NeuroDataWithoutBorders)'};
+ options.dispStr = {'Download Fiji (to run Miji)','Download CNMF, CNMF-E, and CVX code.','Download test one- and two photon datasets.','Load Fiji/Miji into MATLAB path.','Download NWB (NeuroDataWithoutBorders)','Download EXTRACT'};
% Int vector: index of options.dependencyStr to run by default with no GUI
- options.depIdxArray = [1 2 3 5];
+ options.depIdxArray = [1 2 3 5 6];
% get options
options = getOptions(options,varargin);
% display(options)
diff --git a/@calciumImagingAnalysis/modelAddNewFolders.m b/@calciumImagingAnalysis/modelAddNewFolders.m
index 6ca816e..40a8024 100644
--- a/@calciumImagingAnalysis/modelAddNewFolders.m
+++ b/@calciumImagingAnalysis/modelAddNewFolders.m
@@ -26,7 +26,7 @@
try
nExistingFolders = length(obj.inputFolders);
if isempty(options.folderCellArray)
- sel = 0
+ sel = 0;
if isempty(options.inputMethod)
usrIdxChoiceStr = {...
'manually enter folders to list',...
diff --git a/@calciumImagingAnalysis/modelExtractSignalsFromMovie.m b/@calciumImagingAnalysis/modelExtractSignalsFromMovie.m
index 104788e..d526829 100644
--- a/@calciumImagingAnalysis/modelExtractSignalsFromMovie.m
+++ b/@calciumImagingAnalysis/modelExtractSignalsFromMovie.m
@@ -21,6 +21,8 @@
% 2019.11.10 [20:34:42] - Add a warning with some common tips for users if error during cell extraction. Skip modelVarsFromFiles and viewObjmaps loading to reduce user confusion for any folders that had issues during cell extraction.
% 2020.05.08 [20:01:52] - Make creation of settings an explicit option that the user can change.
% 2021.02.01 [15:19:40] - Update `_external_programs` to call ciapkg.getDirExternalPrograms() to standardize call across all functions.
+ % 2021.02.25 [16:44:41] - Update `saveRunTimes` to handle case in which user selects multiple movies for cell extraction.
+ % 2021.03.20 [19:23:25] - Convert ndSparse outputs to single from cell-extraction algorithms (e.g. for CELLMax/EXTRACT) when saving as NWB. Updated EXTRACT support to include additional options.
% TODO
%
@@ -85,7 +87,7 @@
% getAlgorithmRootPath('CELLMax_Wrapper.m','CELLMax',obj);
getAlgorithmRootPath('runCELLMax.m','CELLMax',obj,1);
case 'EXTRACT'
- getAlgorithmRootPath('extractor.m','EXTRACT',obj,1);
+ getAlgorithmRootPath('extractor.m','EXTRACT',obj,0);
otherwise
end
end
@@ -458,7 +460,11 @@ function subfxnSaveNwbFiles(inputImages,inputTraces)
nwbSavePath = [tmpDirHere filesep thisDirSaveStrFile obj.extractionMethodSaveStr.(obj.signalExtractionMethod) '.nwb'];
end
nwbOpts.fpathYML = [obj.externalProgramsDir filesep 'nwb_schnitzer_lab' filesep 'ExampleMetadata.yml'];
- [success] = saveNeurodataWithoutBorders(inputImages,inputTraces,obj.signalExtractionMethod,nwbSavePath,'options',nwbOpts);
+ if issparse(inputImages)
+ [success] = saveNeurodataWithoutBorders(single(full(inputImages)),inputTraces,obj.signalExtractionMethod,nwbSavePath,'options',nwbOpts);
+ else
+ [success] = saveNeurodataWithoutBorders(inputImages,inputTraces,obj.signalExtractionMethod,nwbSavePath,'options',nwbOpts);
+ end
end
end
function getAlgorithmRootPath(algorithmFile,algorithmName,obj,rootFlag)
@@ -470,6 +476,8 @@ function getAlgorithmRootPath(algorithmFile,algorithmName,obj,rootFlag)
if rootFlag==1
[pathToAdd,~,~] = fileparts(pathToAdd);
end
+ % Get all sub-directories
+ pathToAdd = genpath(pathToAdd);
try
pathList = strjoin(pathToAdd,pathsep);
catch
@@ -586,9 +594,17 @@ function saveRunTimes(algorithm)
runtimeTable.daytime{addRow,1} = datestr(now,'HH:MM','local');
runtimeTable.folder{addRow,1} = obj.folderBaseSaveStr{obj.fileNum};
runtimeTable.algorithm{addRow,1} = algorithm;
- runtimeTable.frames(addRow,1) = movieDims.z;
- runtimeTable.width(addRow,1) = movieDims.x;
- runtimeTable.height(addRow,1) = movieDims.y;
+
+ if iscell(movieList)
+ if length(movieList)>1
+ disp("Only adding the first movie's information to the runtime output.")
+ end
+ elseif length(movieDims.z)>1
+ disp("Only adding the first movie's information to the runtime output.")
+ end
+ runtimeTable.frames(addRow,1) = movieDims.z(1);
+ runtimeTable.width(addRow,1) = movieDims.x(1);
+ runtimeTable.height(addRow,1) = movieDims.y(1);
fprintf('saving %s\n',runtimeTablePath);
writetable(runtimeTable,runtimeTablePath,'FileType','text','Delimiter',',');
@@ -831,17 +847,21 @@ function getAlgorithmOptions()
'EXTRACT | Use GPU (''gpu'') or CPU (''cpu'')?',...
'EXTRACT | avg_cell_radius (Avg. cell radius, also controls cell elimination)? Leave blank for GUI to estimate radius.',...
'EXTRACT | cellfind_min_snr (threshold on the max instantaneous per-pixel SNR in the movie for searching for cells)?',...
- 'EXTRACT | preprocess? (1 = yes, 0 = no)',...
+ 'EXTRACT | preprocess? (1 = yes, 0 = no, [ONLY SELECT 1 on raw, motion-corrected movies])',...
'EXTRACT | num_partitions? Int: number of partitions in x and y.',...
- 'EXTRACT | compact_output? (1 = yes, 0 = no)'...
+ 'EXTRACT | compact_output? (1 = yes, 0 = no)',...
+ 'EXTRACT | trace_output_option? Trace output type("nonneg" or "raw")',...
+ 'EXTRACT | use_sparse_arrays? (1 = yes, 0 = no)'...
},...
dlgStr,1,...
{...
'cpu',...
'',...
- '2'...
+ '1'...
'0',...
- '1',...
+ '2',...
+ '0',...
+ 'nonneg',...
'0',...
}...
);setNo = 1;
@@ -852,6 +872,8 @@ function getAlgorithmOptions()
options.EXTRACT.num_partitions_x = str2num(movieSettings{setNo});setNo = setNo+1;
options.EXTRACT.num_partitions_y = options.EXTRACT.num_partitions_x;
options.EXTRACT.compact_output = str2num(movieSettings{setNo});setNo = setNo+1;
+ options.EXTRACT.trace_output_option = movieSettings{setNo};setNo = setNo+1;
+ options.EXTRACT.use_sparse_arrays = str2num(movieSettings{setNo});setNo = setNo+1;
case 'CNMF'
movieSettings = inputdlg({...
@@ -1421,6 +1443,9 @@ function runPCAICASignalFinder()
% end
% extractConfig.num_estimated_cells = nPCsnICs(1);
+ % Load default configuration
+ extractConfig = get_defaults([]);
+
extractConfig.avg_cell_radius = gridWidth.(obj.subjectStr{obj.fileNum});
extractConfig.preprocess = options.EXTRACT.preprocess;
switch options.EXTRACT.gpuOrCPU
@@ -1432,12 +1457,14 @@ function runPCAICASignalFinder()
otherwise
% body
end
- extractConfig.remove_static_background = false;
- extractConfig.skip_dff = true;
-
- % extractConfig.cellfind_min_snr = options.EXTRACT.cellfind_min_snr;
- % extractConfig.num_partitions_x = options.EXTRACT.num_partitions_x;
- % extractConfig.num_partitions_y = options.EXTRACT.num_partitions_y;
+ % extractConfig.remove_static_background = false;
+ % extractConfig.skip_dff = true;
+
+ extractConfig.cellfind_min_snr = options.EXTRACT.cellfind_min_snr;
+ extractConfig.num_partitions_x = options.EXTRACT.num_partitions_x;
+ extractConfig.num_partitions_y = options.EXTRACT.num_partitions_y;
+ extractConfig.trace_output_option = options.EXTRACT.trace_output_option;
+ extractConfig.use_sparse_arrays = options.EXTRACT.use_sparse_arrays;
% extractConfig.compact_output = options.EXTRACT.compact_output;
% extractConfig.thresholds.T_min_snr = 3; % multiply with noise_std
@@ -1463,18 +1490,24 @@ function runPCAICASignalFinder()
extractAnalysisOutput.filters = outStruct.spatial_weights;
% permute so it is [nCells frames]
extractAnalysisOutput.traces = permute(outStruct.temporal_weights, [2 1]);
- extractAnalysisOutput.info = outStruct.info;
- extractAnalysisOutput.config = outStruct.config;
- extractAnalysisOutput.info = outStruct.info;
- % Remove the large summary field since takes up unnecessary space
- extractAnalysisOutput.info.summary = [];
- extractAnalysisOutput.file = movieList{1};
- extractAnalysisOutput.userInputConfig = extractConfig;
- % for backwards compatibility
- extractAnalysisOutput.opts = outStruct.config;
- extractAnalysisOutput.time.startTime = startTime;
- extractAnalysisOutput.time.endTime = tic;
- extractAnalysisOutput.time.totalTime = toc(startTime);
+ try
+ extractAnalysisOutput.info = outStruct.info;
+ extractAnalysisOutput.config = outStruct.config;
+ extractAnalysisOutput.info = outStruct.info;
+ % Remove the large summary field since takes up unnecessary space
+ extractAnalysisOutput.info.summary = [];
+ extractAnalysisOutput.file = movieList{1};
+ extractAnalysisOutput.userInputConfig = extractConfig;
+ % for backwards compatibility
+ extractAnalysisOutput.opts = outStruct.config;
+ extractAnalysisOutput.time.startTime = startTime;
+ extractAnalysisOutput.time.endTime = tic;
+ extractAnalysisOutput.time.totalTime = toc(startTime);
+ catch err
+ display(repmat('@',1,7))
+ disp(getReport(err,'extended','hyperlinks','on'));
+ display(repmat('@',1,7))
+ end
% =======
% save EXTRACT signals
@@ -1852,6 +1885,22 @@ function runPCAICASignalFinder()
end
end
+ if strcmp(signalExtractionMethod{signalExtractNo},'EXTRACT')
+ if ~isempty(options.EXTRACT.avg_cell_radius)
+ display('Use manually entered values.')
+ for thisSubjectStr=subjectList
+ display(repmat('=',1,21))
+ thisSubjectStr = thisSubjectStr{1};
+ display(thisSubjectStr);
+ gridWidth.(thisSubjectStr) = options.EXTRACT.avg_cell_radius;
+ gridSpacing.(thisSubjectStr) = options.EXTRACT.avg_cell_radius;
+ end
+ gridWidth
+ gridSpacing
+ return;
+ end
+ end
+
for thisSubjectStr=subjectList
try
display(repmat('=',1,21))
diff --git a/@calciumImagingAnalysis/runPipeline.m b/@calciumImagingAnalysis/runPipeline.m
index c5709c6..e47337f 100644
--- a/@calciumImagingAnalysis/runPipeline.m
+++ b/@calciumImagingAnalysis/runPipeline.m
@@ -77,7 +77,7 @@
currentIdx = intersect(currentIdx,duplicateIdx);
% [idNumIdxArray, ok] = listdlg('ListString',fxnsToRun,'InitialValue',currentIdx(1),'ListSize',dlgSize,'Name','Sir! I have a plan! Select a calcium imaging analysis method or procedure to run:');
- [idNumIdxArray, fileIdxArray, ok] = obj.calciumImagingAnalysisMainGui(fxnsToRun,['"Sir! I have a plan!" Hover over methods for tooltip descriptions.'],currentIdx);
+ [idNumIdxArray, fileIdxArray, ok] = obj.ciatahMainGui(fxnsToRun,['"Sir! I have a plan!" Hover over methods for tooltip descriptions.'],currentIdx);
obj.foldersToAnalyze = fileIdxArray;
bypassUI = 1;
diff --git a/README.md b/README.md
index de5d102..6396267 100644
--- a/README.md
+++ b/README.md
@@ -30,7 +30,7 @@
- Includes all major calcium imaging analysis steps: movie visualization (including reading from disk), pre-processing (motion correction, spatiotemporal downsampling, spatial filtering, relative fluorescence calculation, etc.), support for multiple cell-extraction methods (CELLMax, PCA-ICA, CNMF, CNMF-E, EXTRACT, etc.), manual classification via GUIs, automated cell classification (coming soon!), cross-session cell alignment, and more.
- Has several example one- and two-photon calcium imaging datasets that `ciatah` can automatically download to help users test out the package.
- Includes code for determining animal position (e.g. in open-field assay).
-- Supports [Neurodata Without Borders](https://www.nwb.org/) data standard (see [calcium imaging tutorial](https://neurodatawithoutborders.github.io/matnwb/tutorials/html/ophys.html)) for reading/writing cell-extraction (e.g. outputs of PCA-ICA, CELLMax, CNMF, CNMF-E, etc.). Supports reading and writing NWB movie files with continued integration planned.
+- Supports [Neurodata Without Borders](https://www.nwb.org/) data standard (see [calcium imaging tutorial](https://neurodatawithoutborders.github.io/matnwb/tutorials/html/ophys.html)) for reading/writing cell-extraction (e.g. outputs of PCA-ICA, CELLMax, CNMF, CNMF-E, EXTRACT, etc.). Supports reading and writing NWB movie files with continued integration planned.
- Supports most major imaging movie file formats: HDF5, NWB, AVI, ISXD [Inscopix], and TIFF.
- Requires `MATLAB`.
@@ -251,7 +251,7 @@ Please email any additional questions not covered in the repository to `github [
## License
-Copyright (C) 2013-2020 Biafra Ahanonu
+Copyright (C) 2013-2021 Biafra Ahanonu
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
diff --git a/ciapkg/VERSION b/ciapkg/VERSION
index fa8dfe1..7f68407 100644
--- a/ciapkg/VERSION
+++ b/ciapkg/VERSION
@@ -1,2 +1,2 @@
-v3.24.1
-20210221190557
\ No newline at end of file
+v3.25.1
+20210320182424
\ No newline at end of file
diff --git a/ciapkg/classification/signalSorter.m b/ciapkg/classification/signalSorter.m
index 0719769..7b5fb16 100644
--- a/ciapkg/classification/signalSorter.m
+++ b/ciapkg/classification/signalSorter.m
@@ -70,6 +70,7 @@
% 2020.09.22 [03:09:59] - Ensure NWB files are read correctly.
% 2020.10.13 [01:22:47] - Display context menu for keyboard shortcuts, easier than separate figure. User can select with right-click or via a menu in the GUI.
% 2020.10.13 [22:31:23] - Users can now scroll through cells using mouse scroll wheel.
+ % 2021.03.10 [16:33:23] - User can now input just NWB path without a blank variable for inputSignals. Also added support for CIAtah mat files.
% TODO
% DONE: New GUI interface to allow users to scroll through video and see cell activity at that point
% DONE: allow option to mark rest as bad signals
@@ -241,10 +242,19 @@
end
display(options)
- % If inputs NWB format
- if ischar(inputImages)
- [inputImages,inputSignals,infoStruct] = loadNeurodataWithoutBorders(inputImages);
- options.nSignals = size(inputImages,3);
+ % If inputs are NWB or CIAtah mat format
+ if nargin==1|ischar(inputImages)
+ [~,~,inputEXT] = fileparts(inputImages);
+ switch inputEXT
+ case '.nwb'
+ [inputImages,inputSignals,infoStruct,algorithmStr] = loadNeurodataWithoutBorders(inputImages);
+ options.nSignals = size(inputImages,3);
+ case '.mat'
+ [inputImages,inputSignals,infoStruct,algorithmStr] = ciapkg.io.loadSignalExtraction(inputImages);
+ options.nSignals = size(inputImages,3);
+ otherwise
+
+ end
end
% Modify dataset name if given NWB file
diff --git a/ciapkg/io/getFileInfo.m b/ciapkg/io/getFileInfo.m
index 96a28b6..6d43692 100644
--- a/ciapkg/io/getFileInfo.m
+++ b/ciapkg/io/getFileInfo.m
@@ -28,7 +28,8 @@
'|vehicle_ldopa|vehicle_skf|vehicle_quinpirole',...
'|habita|habitb|fc|contextrecall|recall|renewal',...
'|unc9975|unc|ari|hal|ketamine|unca\d+_unca|prePD|postPD|activeAvoidance|passiveAvoidance|baseline',...
- '|sncEighty|ram|sl|set|D'};
+ '|sncEighty|ram|sl|set|D',...
+ '|epi|2p'};
% second list of assays to search for
% options.assayListPre = {...
% 'veh(|icle)_unc\d+_amph|veh(|icle)_ari\d+_amph|veh(|icle)_hal\d+_amph',...
diff --git a/ciapkg/io/loadNeurodataWithoutBorders.m b/ciapkg/io/loadNeurodataWithoutBorders.m
index 6155355..160bedf 100644
--- a/ciapkg/io/loadNeurodataWithoutBorders.m
+++ b/ciapkg/io/loadNeurodataWithoutBorders.m
@@ -5,7 +5,10 @@
% inputs
% inputFilePath - Str: path to NWB file. If a cell, will only load the first string.
% outputs
- %
+ % inputImages - 3D or 4D matrix containing cells and their spatial information, format: [x y nCells].
+ % inputSignals - 2D matrix containing activity traces in [nCells nFrames] format.
+ % infoStruct - contains information about the file, e.g. the 'description' property that can contain information about the algorithm.
+ % algorithmStr - String of the algorithm name.
% changelog
% 2021.02.05 [19:02:44] - Parse the algorithm associated with the NWB signal extraction data.
diff --git a/ciapkg/io/saveNeurodataWithoutBorders.m b/ciapkg/io/saveNeurodataWithoutBorders.m
index 145eae0..87171e4 100644
--- a/ciapkg/io/saveNeurodataWithoutBorders.m
+++ b/ciapkg/io/saveNeurodataWithoutBorders.m
@@ -14,9 +14,10 @@
% changelog
% 2020.07.01 [09:40:20] - Convert roi_response_data to cell if user inputs only a matrix.
% 2020.09.15 [20:30:32] - Automatically creates directory where file is to be stored if it is not present.
- % 2021.02.01 [�?15:14:40] - Function checks that yaml, matnwb, and nwb_schnitzer_lab loaded, else tries to load to make sure all dependencies are present and active.
+ % 2021.02.01 [?15:14:40] - Function checks that yaml, matnwb, and nwb_schnitzer_lab loaded, else tries to load to make sure all dependencies are present and active.
% 2021.02.01 [15:19:40] - Update `_external_programs` to call ciapkg.getDirExternalPrograms() to standardize call across all functions.
% 2021.02.03 [12:34:06] - Added a check for inputs with a single signal and function returns as it is not supported.
+ % 2021.03.20 [19:35:28] - Update to checking if only a single signal input.
% TODO
%
@@ -105,8 +106,10 @@
tmpData = roi_response_data;
roi_response_data = struct;
+ disp(['Input images size: ' num2str(size(image_masks))])
for i=1:length(tmpData)
- if size(1,tmpData{i})==1
+ disp(['Input traces #' num2str(i) ' size: ' num2str(size(tmpData{i}))])
+ if size(tmpData{i},1)==1
disp('Only a single output, NWB will not support at the moment.')
return;
end
diff --git a/ciapkg/signal_extraction/computeCnmfeSignalExtraction_batch.m b/ciapkg/signal_extraction/computeCnmfeSignalExtraction_batch.m
index 2dfaa51..ae3247f 100644
--- a/ciapkg/signal_extraction/computeCnmfeSignalExtraction_batch.m
+++ b/ciapkg/signal_extraction/computeCnmfeSignalExtraction_batch.m
@@ -19,6 +19,7 @@
% changelog
% 2016.06.20 - updated to keep in line with recent changes to CNMF functions
% 2021.01.24 [14:29:06] - Added trace origin type to output structure.
+ % 2021.03.20 [20:20:27] - extractedSignalsType, extractedSignalsEstType struct update.
% TODO
%
@@ -386,8 +387,8 @@
cnmfeAnalysisOutput.extractedImages = reshape(full(results.A),[neuron.options.d1 neuron.options.d2 size(results.C,1)]);
cnmfeAnalysisOutput.extractedSignals = results.C;
cnmfeAnalysisOutput.extractedSignalsEst = results.C_raw;
- cnmfAnalysisOutput.extractedSignalsType = 'model';
- cnmfAnalysisOutput.extractedSignalsEstType = 'dfof';
+ cnmfeAnalysisOutput.extractedSignalsType = 'model';
+ cnmfeAnalysisOutput.extractedSignalsEstType = 'dfof';
cnmfeAnalysisOutput.extractedPeaks = results.S;
cnmfeAnalysisOutput.Cn = results.Cn;
cnmfeAnalysisOutput.P = results.P;
diff --git a/ciapkg/view/changeFont.m b/ciapkg/view/changeFont.m
index a903814..def8279 100644
--- a/ciapkg/view/changeFont.m
+++ b/ciapkg/view/changeFont.m
@@ -10,6 +10,7 @@
% changelog
% 2020.04.28 [19:17:49] - Added ability to change all the font colors at the same time, useful for making presentations on non-white backgrounds.
% 2020.10.01 [09:39:33] - Added support for changing font type.
+ % 2021.03.07 [16:35:55] - Add font name support.
% TODO
% Add support for changing other font aspects, e.g. figure Font family, command window font, etc.
@@ -19,6 +20,8 @@
options.FontSize = [];
% Float: [r g b] vector between 0 to 1.
options.fontColor = [];
+ % Str: Name of font family, e.g. Consolas.
+ options.fontName = [];
% get options
options = getOptions(options,varargin);
% display(options)
@@ -50,7 +53,11 @@
catch
end
- end
+ end
+ if ~isempty(options.fontName)
+ set(findall(gcf,'-property','FontName'),'FontName',options.fontName);
+ end
+
end
success = 1;
catch err
diff --git a/docs/README.md b/docs/README.md
index cdaa5f0..b456670 100644
--- a/docs/README.md
+++ b/docs/README.md
@@ -1,6 +1,6 @@
-# calciumImagingAnalysis documentation
+# CIAtah documentation
-This directory contains the documentation for `calciumImagingAnalysis` that can be built from the Markdown files using `Python 3.7` or greater.
+This directory contains the documentation for `CIAtah` that can be built from the Markdown files using `Python 3.7` or greater.
## Instructions
@@ -8,4 +8,16 @@ The documentation can always be found at https://bahanonu.github.io/calciumImagi
Run `make_docs_local.py`, e.g. `python -m make_docs_local.py` in the command line, to make the documents to view locally.
-Run `make_docs.py`, e.g. `python -m make_docs.py` in the command line, to make the documents for online, e.g. where each page is a directory with a `index.html` file.
\ No newline at end of file
+Run `make_docs.py`, e.g. `python -m make_docs.py` in the command line, to make the documents for online, e.g. where each page is a directory with a `index.html` file.
+
+## Dependencies
+
+Run the below `pip` commands to make sure all dependencies are installed.
+
+```
+pip install mkdocs
+
+pip install mdx_truly_sane_lists
+
+pip install pymdown-extensions
+```
\ No newline at end of file
diff --git a/docs/docs/css/extra.css b/docs/docs/css/extra.css
index aa61a39..992e1ec 100644
--- a/docs/docs/css/extra.css
+++ b/docs/docs/css/extra.css
@@ -34,4 +34,116 @@ html{
overflow-wrap: break-word;
word-wrap: break-word;
background-color: black;
-}
\ No newline at end of file
+}
+.codehilite pre code{
+ /*overflow-x: auto;
+ white-space: pre-wrap;
+ white-space: -moz-pre-wrap;
+ white-space: -pre-wrap;
+ white-space: -o-pre-wrap;
+ word-wrap: break-word;*/
+ /*background-color: black;*/
+}
+/*html{
+ background-color: black;
+}
+pre code {
+ white-space: pre-wrap;
+}
+code:before {
+ counter-reset: listing;
+}
+pre.code code {
+ counter-increment: listing;
+}
+pre.code code::before {
+ content: counter(listing) ". ";
+ display: inline-block;
+ width: 8em;
+ padding-left: auto;
+ margin-left: auto;
+ text-align: right;
+}*/
+.highlight pre{
+ padding: 0;
+}
+.lineno{
+ color: gray;
+ font-style: italic;
+}
+
+[data-linenos]:before {
+ content: attr(data-linenos);
+ /*padding-right: 10px;*/
+ /*display: table-cell;*/
+}
+h2{
+ padding-top: 1em;
+ border-top: 3px solid black;
+}
+/*h2:before{*/
+ /*content:
;*/
+/*}*/
+hr{
+ height: 0px;
+ border-top: 3px solid black;
+}
+.caption{
+ border-top: 3px solid #333300;
+}
+.wy-menu-vertical li.current{
+}
+.wy-menu-vertical li.toctree-l1.current>a{
+ background-color: #161616;
+ color: #d9d9d9;
+ border-top: none;
+ border-bottom: none;
+ /*solid 1px #c9c9c9*/
+}
+.wy-menu-vertical li.toctree-l1{
+ background-color: black;
+}
+.wy-menu-vertical li.toctree-l1 a{
+ color: #d9d9d9;
+}
+
+.wy-menu-vertical li.toctree-l2{
+ background-color: black;
+}
+.wy-menu-vertical li.toctree-l2 a{
+ color: #d9d9d9;
+}
+
+
+.wy-menu-vertical li.toctree-l2.current>a{
+ background-color: #161616;
+ color: #d9d9d9;
+}
+
+.wy-menu-vertical li.toctree-l2.current li.toctree-l3>a{
+ background-color: black;
+ color: #d9d9d9;
+}
+
+.wy-menu-vertical li.toctree-l3{
+ background-color: black;
+}
+.wy-menu-vertical li.toctree-l3 a{
+ color: #d9d9d9;
+}
+
+.wy-menu-vertical li.toctree-l2 a:hover{
+ background-color: #161616;
+}
+
+.wy-side-nav-search{
+ background-color: black;
+}
+
+.subsection1{
+ border-bottom: 3px solid #333300;
+ padding-top: 1em;
+ font-weight: bold;
+ /*background-color: #161616;*/
+ /*color: #d9d9d9;*/
+}
diff --git a/docs/docs/data.md b/docs/docs/data.md
index 4393031..61b9a25 100644
--- a/docs/docs/data.md
+++ b/docs/docs/data.md
@@ -7,15 +7,23 @@ The naming convention in general is below. Both TIF and AVI raw files are suppor
### Input and output files
- Default raw imaging data filename: `concat_.*.(h5|tif)`.
- Default raw processed data filename: `folderName_(processing steps).h5`, where `folderName` is the directory name where the calcium imaging movies are located.
-- Main files output by `calciumImagingAnalysis`. Below, `.*` normally indicates the folder name prefixed to the filename.
+- Main files output by `{{ site.name }}`. Below, `.*` normally indicates the folder name prefixed to the filename.
- `.*_pcaicaAnalysis.mat`: Where PCA-ICA outputs are stored.
- `.*_ICdecisions_.*.mat`: Where decisions for cell (=1) and not cell (=0) are stored in a `valid` variable.
- `.*_regionModSelectUser.mat`: A mask of the region (=1) to include in further analyses.
- `.*_turboreg_crop_dfof_1.h5`: Processed movie, in this case motion corrected, cropped, and Δ_F/F_.
- `processing_info`: a folder containing preprocessing information.
+### Loading data
+
+Users can load data from any NWB or {{ site.name }}-style MAT files containing cell-extraction outputs using the `ciapkg.io.loadSignalExtraction` function as below.
+
+```Matlab
+[inputImages,inputSignals,infoStruct,algorithmStr,inputSignals2] = ciapkg.io.loadSignalExtraction(fileName);
+```
+
### NWB Support
-calciumImagingAnalysis supports NWB format and by default will output cell-extraction analysis as calciumImagingAnalysis format unless user specifies otherwise. NWB files are by default stored in the `nwbFiles` sub-folder. This can be changed by setting the `obj.nwbFileFolder` property to a different folder name.
+{{ site.name }} supports NWB format and by default will output cell-extraction analysis as {{ site.name }} format unless user specifies otherwise. NWB files are by default stored in the `nwbFiles` sub-folder. This can be changed by setting the `obj.nwbFileFolder` property to a different folder name. Learn more about saving and loading
- Default image mask HDF5 dataset name: `/processing/ophys/ImageSegmentation/PlaneSegmentation`.
- Default fluorescence activity HDF5 dataset name: `/processing/ophys/Fluorescence/RoiResponseSeries`.
diff --git a/docs/docs/help_spatial_filtering.md b/docs/docs/help_spatial_filtering.md
index f192de5..801baa2 100644
--- a/docs/docs/help_spatial_filtering.md
+++ b/docs/docs/help_spatial_filtering.md
@@ -1,28 +1,69 @@
# Movie Filtering
-This page documents different functions in the repository variable for filtering (spatial high/low/bandpass) movies to remove neuropil, cells, or other features.
+This page documents different features and functions in the {{ site.name }} repository variable for filtering (spatial high/low/bandpass) movies to remove neuropil, cells, or other features.
-###
+
-```Matlab
+
+## Filtering movies with `normalizeMovie`
+
+Users can quickly filter movies using the `normalizeMovie` function. See below for usage.
+
+```Matlab
+% Load the movie (be in TIF, HDF5, AVI, etc.). Change HDF5 input dataset name as needed.
+inputMovie = loadMovieList('pathToMovie','inputDatasetName','/1');
+
+% Set options for dividing movie by lowpass version of the movie
options.freqLow = 1;
options.freqHigh = 4;
-inputMovie = normalizeMovie(single(inputMovie),'normalizationType','lowpassFFTDivisive','freqLow',options.freqLow,'freqHigh',options.freqHigh,'waitbarOn',1,'bandpassMask','gaussian');
+options.normalizationType = 'lowpassFFTDivisive';
+options.waitbarOn = 1;
+options.bandpassMask = 'gaussian';
+
+% Options for normal bandpass filter
+options.freqLow = 10;
+options.freqHigh = 50;
+options.normalizationType = 'fft';
+options.waitbarOn = 1;
+options.bandpassMask = 'gaussian';
+
+% Set additional common options
+options.showImages = 0; % Set to 1 if you want to view
+
+% Run analysis
+inputMovie = normalizeMovie(single(inputMovie),'options',options);
```
+If users set `options.showImages = 0;`, then `normalizeMovie` will update a figure containing both real and frequency space before and after the filter has been applied along with an example of the filter in frequency space. This allows users to get a sense of what their filter is doing.
+
+
+
+## Why conduct spatial filtering?
+
+Spatial filtering can have a large impact on the resulting cell activity traces extracted from the movies.
+
+For example, below are the correlations between all cell-extraction outputs from PCA-ICA, ROI back-application of ICA filters, and CNMF-e on a miniature microscope one-photon movie. As can be seen, especially in the case of ROI analysis, the correlation between the activity traces is rendered artificially high due to the correlated background noise. This is greatly reduced in many instances after proper spatial filtering.
+
+
+
+
+
+
+
## Images from unit test
### Main filtering functions.
Below is a screen grab from a random frame using all the filtering functions. A nice way to quickly see the many differences between each functions filtering.

### Test function filtering
-This is currently only for the Matlab fft, but I'll see about expanding to others.
-
+This function will take a movie and This is currently only for the Matlab fft, but I'll see about expanding to others.
```Matlab
unitNormalizeMovie;
```
+
+
### Matlab test function
* I've also added the ability to test the parameter space of the Matlab fft, use the below command.
```Matlab
@@ -33,7 +74,7 @@ testMovieFFT = normalizeMovie(testMovie,'normalizationType','matlabFFT_test','se
### Matlab test function movie output
-Similar to above, showing results when using `lowpassFFTDivisive` normalization (`matlab divide by lowpass before registering` in `modelPreprocessMovie` and `viewMovieRegistrationTest` functions) with `freqLow = 0` and `freqHigh` set to `1`, `4`, and `20`. This corresponds to removing increasingly smaller features from the movie.
+Similar to above, showing results when using `lowpassFFTDivisive` normalization (using the `matlab divide by lowpass before registering` option in `modelPreprocessMovie` and `viewMovieRegistrationTest` functions) with `freqLow = 0` and `freqHigh` set to `1`, `4`, and `20`. This corresponds to removing increasingly smaller features from the movie.

@@ -46,6 +87,8 @@ inputMovieTest = normalizeMovie(inputMovie,'normalizationType','imagejFFT_test')
The output should look like the below:

+
+
## Common Issues
A list of some common issues.
@@ -74,6 +117,7 @@ testMovieFFT = normalizeMovie(testMovie,'normalizationType','lowpassFFTDivisive'
testMovieFFTImageJ = normalizeMovie(testMovie,'normalizationType','imagejFFT');
diffMovie = testMovieFFT-testMovieFFTImageJ ;
```
+
* With some tweaking of the `freqHigh` and other parameters, should hopefully be able to get closer to macheps and say that the two are identical for our purposes.

diff --git a/docs/docs/index.md b/docs/docs/index.md
index 3b4f92f..1d43a9e 100644
--- a/docs/docs/index.md
+++ b/docs/docs/index.md
@@ -1,4 +1,4 @@
-# CIAtah (calciumImagingAnalysis [`CIAPKG`])
+# {{ site.name }} (calciumImagingAnalysis [`CIAPKG`])


[](https://github.com/bahanonu/calciumImagingAnalysis/releases/latest?style=flat-square&logo=appveyor)
@@ -11,20 +11,20 @@
Created by __Biafra Ahanonu, PhD__.
-`CIAtah` (pronounced cheetah) or `calciumImagingAnalysis` (`CIAPKG`) is a software package for analysis of one- and two-photon calcium imaging datasets.
+`{{ site.name }}` (pronounced cheetah) or `calciumImagingAnalysis` (`CIAPKG`) is a software package for analysis of one- and two-photon calcium imaging datasets.
__Download the software at [https://github.com/bahanonu/calciumImagingAnalysis](https://github.com/bahanonu/calciumImagingAnalysis).__
- +
+ -- This user guide contains instructions to setup, run, and troubleshoot `CIAtah`.
-- Note: `CIAtah` is a class (`ciatah` or `calciumImagingAnalysis` within MATLAB) with various GUIs to allow processing of calcium imaging data. In addition, users can access the underlying `CIAtah` functions to make custom workflows. See [Custom command-line pipelines](api_example_pipeline.md).
+- This user guide contains instructions to setup, run, and troubleshoot `{{ site.name }}`.
+- Note: `{{ site.name }}` is a class (`{{ site.namelow }}` or `calciumImagingAnalysis` within MATLAB) with various GUIs to allow processing of calcium imaging data. In addition, users can access the underlying `{{ site.name }}` functions to make custom workflows. See [Custom command-line pipelines](api_example_pipeline.md).
- Read my overview of calcium imaging analysis methods at [Calcium imaging cell identification and fluorescence activity trace reconstruction, part 1](https://bahanonu.com/brain/#c20181209).
-- This user guide contains instructions to setup, run, and troubleshoot `CIAtah`.
-- Note: `CIAtah` is a class (`ciatah` or `calciumImagingAnalysis` within MATLAB) with various GUIs to allow processing of calcium imaging data. In addition, users can access the underlying `CIAtah` functions to make custom workflows. See [Custom command-line pipelines](api_example_pipeline.md).
+- This user guide contains instructions to setup, run, and troubleshoot `{{ site.name }}`.
+- Note: `{{ site.name }}` is a class (`{{ site.namelow }}` or `calciumImagingAnalysis` within MATLAB) with various GUIs to allow processing of calcium imaging data. In addition, users can access the underlying `{{ site.name }}` functions to make custom workflows. See [Custom command-line pipelines](api_example_pipeline.md).
- Read my overview of calcium imaging analysis methods at [Calcium imaging cell identification and fluorescence activity trace reconstruction, part 1](https://bahanonu.com/brain/#c20181209).
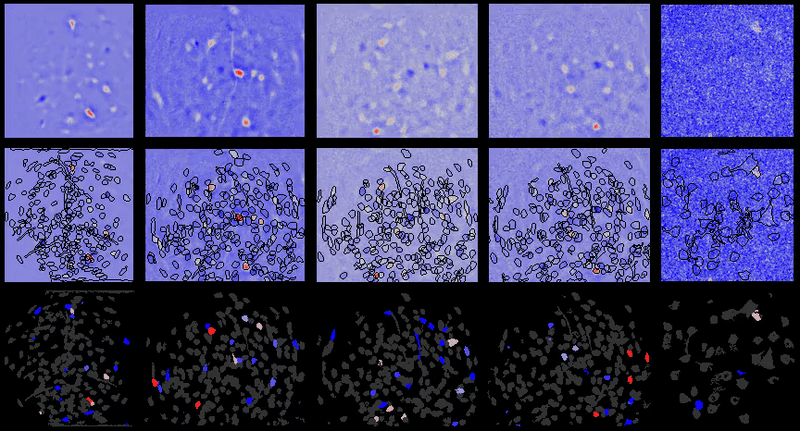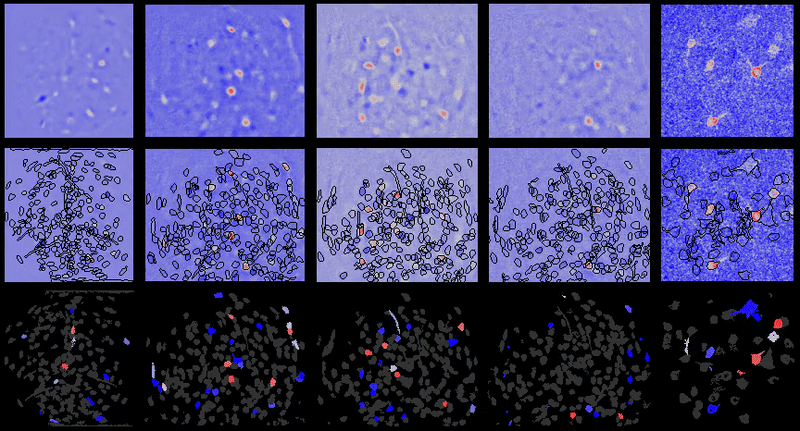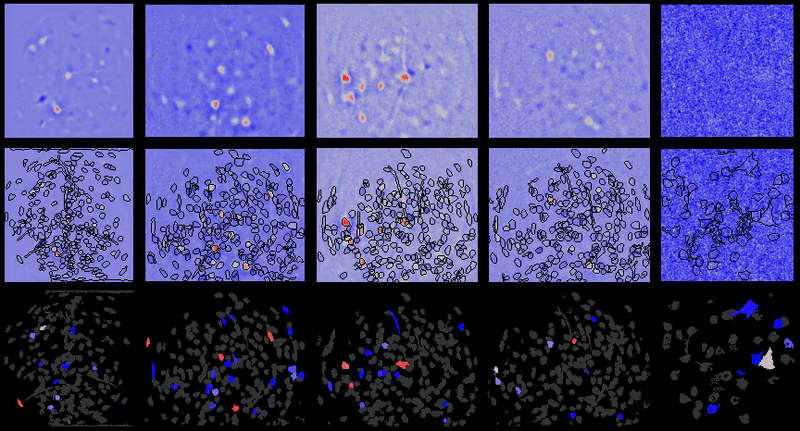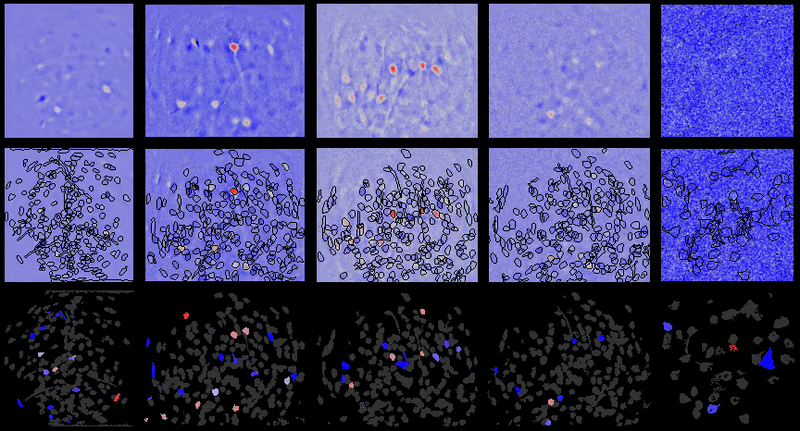
- CIAtah sell sorting GUI
+ {{ site.name }} cell sorting GUI
@@ -32,7 +32,7 @@ __Download the software at [https://github.com/bahanonu/calciumImagingAnalysis](
-`CIAtah` features:
+`{{ site.name }}` features:
- Includes a GUI to allow users to do large-scale batch analysis, accessed via the repository's `calciumImagingAnalysis` class.
- The underlying functions can also be used to create GUI-less, command line-ready analysis pipelines. Functions located in `ciapkg` and `+ciapkg` sub-folders.
@@ -48,10 +48,10 @@ __Download the software at [https://github.com/bahanonu/calciumImagingAnalysis](
The main sections of the site:
-- `Setup` - installation of `CIAtah`.
+- `Setup` - installation of `{{ site.name }}`.
- `Repository` - notes about the software package and data formats.
-- `Processing data` - sections related to processing calcium imaging movies using the `CIAtah` class.
-- `API` - details how to run `CIAtah` from the command line. Will include more details on the many underlying functions in the future.
+- `Processing data` - sections related to processing calcium imaging movies using the `{{ site.name }}` class.
+- `API` - details how to run `{{ site.name }}` from the command line. Will include more details on the many underlying functions in the future.
- `Help` - several section that provide hints and help for processing calcium imaging.
- `Misc` - miscellaneous information about the repository.
diff --git a/docs/docs/pipeline_detailed.md b/docs/docs/pipeline_detailed.md
index a3959d5..1b6da50 100644
--- a/docs/docs/pipeline_detailed.md
+++ b/docs/docs/pipeline_detailed.md
@@ -1,10 +1,16 @@
-# Detailed processing pipeline
+# Detailed {{ site.name }} processing pipeline
+
+The following detailed pipeline assumes you have started a {{ site.name }} object using the below command:
+
+```Matlab
+obj = ciatah;
+```
## Spatially downsample raw movies or convert to HDF5 with `modelDownsampleRawMovies`
Users have the ability to spatially downsample raw movies, often necessary to denoise the data, save storage space, and improve runtimes of later processing steps. For most data, users can downsample 2 or 4 times in each spatial dimension while still retaining sufficient pixels per cell to facilitate cell-extraction.
-To run, either select `modelDownsampleRawMovies` in the GUI menu or type the below command after initializing a calciumImagingAnalysis obj.
+To run, either select `modelDownsampleRawMovies` in the GUI menu or type the below command after initializing a {{ site.name }} obj.
```Matlab
obj.modelDownsampleRawMovies;
@@ -21,7 +27,7 @@ The function will automatically put each file in its corresponding folder, __mak
### Converting Inscopix ISXD files to HDF5
-To convert from Inscopix ISXD file format (output by nVista v3+ and nVoke) to HDF5 run `modelDownsampleRawMovies` without changing the regular expression or make sure it looks for `.*.isxd` or similar. Users will need the latest version of the [Inscopix Data Processing Software](https://www.inscopix.com/nVista#Data_Analysis) as these functions take advantage of their API. If calciumImagingAnalysis cannot automatically find the API, it will ask the user to direct it to the _root_ location of the Inscopix Data Processing Software (see below).
+To convert from Inscopix ISXD file format (output by nVista v3+ and nVoke) to HDF5 run `modelDownsampleRawMovies` without changing the regular expression or make sure it looks for `.*.isxd` or similar. Users will need the latest version of the [Inscopix Data Processing Software](https://www.inscopix.com/nVista#Data_Analysis) as these functions take advantage of their API. If {{ site.name }} cannot automatically find the API, it will ask the user to direct it to the _root_ location of the Inscopix Data Processing Software (see below).

@@ -42,7 +48,7 @@ Within each folder will be a sub-folder called `preprocRunTest` inside of which
## Preprocessing calcium imaging movies with `modelPreprocessMovie`
-After users instantiate an object of the `calciumImagingAnalysis` class and enter a folder, they can start preprocessing of their calcium imaging data with `modelPreprocessMovie`.
+After users instantiate an object of the `{{ site.name }}` class and enter a folder, they can start preprocessing of their calcium imaging data with `modelPreprocessMovie`.
- See below for a series of windows to get started, the options for motion correction, cropping unneeded regions, Δ_F/F_, and temporal downsampling were selected for use in the study associated with this repository.
- If users have not specified the path to Miji, a window appears asking them to select the path to Miji's `scripts` folder.
@@ -75,7 +81,7 @@ The algorithm will then run all the requested preprocessing steps and presented

-******************************************
+
## Manual movie cropping with `modelModifyMovies`
@@ -83,7 +89,7 @@ If users need to eliminate specific regions of their movie before running cell e

-******************************************
+
## Extracting cells with `modelExtractSignalsFromMovie`
@@ -91,7 +97,7 @@ Users can run PCA-ICA, CNMF, CNMF-E, and ROI cell extraction by following the be
We normally estimate the number of PCs and ICs on the high end, manually sort to get an estimate of the number of cells, then run PCA-ICA again with IC 1.5-3x the number of cells and PCs 1-1.5x number of ICs.
-To run CNMF or CNMF-E, run `loadDependencies` module (e.g. `obj.loadDependencies`) after calciumImagingAnalysis class is loaded. CVX (a CNMF dependency) will also be downloaded and `cvx_setup` run to automatically set it up.
+To run CNMF or CNMF-E, run `loadDependencies` module (e.g. `obj.loadDependencies`) after {{ site.name }} class is loaded. CVX (a CNMF dependency) will also be downloaded and `cvx_setup` run to automatically set it up.

@@ -101,20 +107,38 @@ The resulting output (on _Figure 45+_) at the end should look something like:
-******************************************
+
+
+## Loading cell-extraction output data for custom scripts
+
+Users can load outputs from cell extraction using the below command. This will then allow users to use the images and activity traces for downstream analysis as needed.
+
+```Matlab
+[inputImages,inputSignals,infoStruct,algorithmStr,inputSignals2] = ciapkg.io.loadSignalExtraction('pathToFile');
+```
+
+Note, the outputs correspond to the below:
+
+- `inputImages` - 3D or 4D matrix containing cells and their spatial information, format: [x y nCells].
+- `inputSignals` - 2D matrix containing activity traces in [nCells nFrames] format.
+- `infoStruct` - contains information about the file, e.g. the 'description' property that can contain information about the algorithm.
+- `algorithmStr` - String of the algorithm name.
+- `inputSignals2` - same as inputSignals but for secondary traces an algorithm outputs.
+
+
## Loading cell-extraction output data with `modelVarsFromFiles`
-In general, after running cell-extraction (`modelExtractSignalsFromMovie`) on a dataset, run the `modelVarsFromFiles` module. This allows `calciumImagingAnalysis` to load/pre-load information about that cell-extraction run.
+In general, after running cell-extraction (`modelExtractSignalsFromMovie`) on a dataset, run the `modelVarsFromFiles` module. This allows `{{ site.name }}` to load/pre-load information about that cell-extraction run.
-If you had to restart MATLAB or are just loading calciumImagingAnalysis fresh but have previously run cell extraction, run this method before doing anything else with that cell-extraction data.
+If you had to restart MATLAB or are just loading {{ site.name }} fresh but have previously run cell extraction, run this method before doing anything else with that cell-extraction data.
A menu will pop-up like below when `modelVarsFromFiles` is loaded, you can normally just leave the defaults as is.

-******************************************
+
## Validating cell extraction with `viewCellExtractionOnMovie`
@@ -124,10 +148,21 @@ Below is an example, with black outlines indicating location of cell extraction

-******************************************
+
## Sorting cell extraction outputs with `computeManualSortSignals`
-Outputs from PCA-ICA (and most other common cell extraction algorithms like CNMF, etc.) output signal sources that are not cells and thus must be manually removed from the output. The repository contains a GUI for sorting cells from not cells. GUI also contains a shortcut menu that users can access by right-clicking or selecting the top-left menu.
+
+
+ {{ site.name }} cell sorting GUI
+
+
+
+  +
+
+
+
+
+
+Outputs from most common cell-extraction algorithms like PCA-ICA, CNMF, etc. contain signal sources that are not cells and thus must be manually removed from the output. The repository contains a GUI for sorting cells from not cells. GUI also contains a shortcut menu that users can access by right-clicking or selecting the top-left menu.
Below users can see a list of options that are given before running the code, those highlighted in green
@@ -138,11 +173,9 @@ Below users can see a list of options that are given before running the code, th
- To manually sort on large movies that will not fit into RAM, select the below options (highlighted in green). This will load only chunks of the movie asynchronously into the GUI as you sort cell extraction outputs.

-### Usage
+### Cell sorting from the command line with `signalSorter`
-Below are two examples of the interface and code to run if not using the `calciumImagingAnalysis` GUI.
-
-Usage instructions below for `signalSorter.m`:
+Usage instructions below for `signalSorter`, e.g. if not using the `{{ site.name }}` GUI.
__Main inputs__
@@ -171,6 +204,8 @@ iopts.backgroundNeutral = repmat(230,[1 3])/255;
[inputImagesSorted, inputSignalsSorted, choices] = signalSorter(inputImages, inputSignals, 'options',iopts);
```
+Examples of the interface on two different datasets:
+
#### BLA one-photon imaging data signal sorting GUI

@@ -183,7 +218,7 @@ iopts.backgroundNeutral = repmat(230,[1 3])/255;
 -******************************************
+
## Removing cells not within brain region with `modelModifyRegionAnalysis`
@@ -191,7 +226,7 @@ If the imaging field-of-view includes cells from other brain regions, they can b

-******************************************
+
## Cross-session cell alignment with `computeMatchObjBtwnTrials`
@@ -221,4 +256,4 @@ To evaluate how well cross-session alignment works, `computeMatchObjBtwnTrials`
### Save cross-session cell alignment with `modelSaveMatchObjBtwnTrials`
-Users can save out the alignment structure by running `modelSaveMatchObjBtwnTrials`. This will allow users to select a folder where `calciumImagingAnalysis` will save a MAT-file with the alignment structure information for each animal.
\ No newline at end of file
+Users can save out the alignment structure by running `modelSaveMatchObjBtwnTrials`. This will allow users to select a folder where `{{ site.name }}` will save a MAT-file with the alignment structure information for each animal.
\ No newline at end of file
diff --git a/docs/docs/pipeline_overview.md b/docs/docs/pipeline_overview.md
index c56cdbc..076ac0d 100644
--- a/docs/docs/pipeline_overview.md
+++ b/docs/docs/pipeline_overview.md
@@ -5,14 +5,14 @@ The general pipeline for processing calcium imaging data is below. This reposito

-To start using the `calciumImagingAnalysis` software package, enter the following into the MATLAB command window.
+To start using the `{{ site.name }}` software package, enter the following into the MATLAB command window.
```Matlab
% Loads all directories
loadBatchFxns;
% Loads the class into an object.
-obj = calciumImagingAnalysis;
+obj = ciatah;
% Open the class menu
obj % then hit enter, no semicolon!
@@ -23,14 +23,14 @@ obj.runPipeline; % then hit enter!
The general order of functions that users should run is ([optional] are those not critical for most datasets):
- `loadDependencies`
- - If user is running calciumImagingAnalysis for the first time, this module has several options to download and load CNMF/CNMF-E code for cell extraction, Fiji for viewing/modifying videos (using Miji), and test data from a miniature microscope experiment.
+ - If user is running {{ site.name }} for the first time, this module has several options to download and load CNMF/CNMF-E code for cell extraction, Fiji for viewing/modifying videos (using Miji), and test data from a miniature microscope experiment.
- `modelDownsampleRawMovies` [optional]
- If users have raw calcium imaging data that needs to be spatially downsampled, e.g. raw data from Inscopix nVista software.
- `modelAddNewFolders`
- Users should always use this method first, used to add folders to the current class object.
- - For example, if users ran `example_downloadTestData.m`, then add the folder `[githubRepoPath]\data\2014_04_01_p203_m19_check01_raw` where `githubRepoPath` is the absolute path to the current `calciumImagingAnalysis` repository.
+ - For example, if users ran `example_downloadTestData.m`, then add the folder `[githubRepoPath]\data\2014_04_01_p203_m19_check01_raw` where `githubRepoPath` is the absolute path to the current `{{ site.name }}` repository.
- `viewMovie`
- - Users should check that calciumImagingAnalysis loads their movies correctly and that Miji is working.
+ - Users should check that {{ site.name }} loads their movies correctly and that Miji is working.
- Users can view movies from disk, which allows checking of very large movies quickly.
- Remember to check that `Imaging movie regexp:` (regular expression class uses to find user movies within given folders) setting matches name of movies currently in repository.
- `viewMovieRegistrationTest` [optional]
@@ -38,7 +38,7 @@ The general order of functions that users should run is ([optional] are those no
- `tregRunX` folders (where `X` is a number) contain details of each run setting. Delete from analysis folder if don't need outputs later.
- Remember to adjust contrast in resulting montage movies since different filtering will change the absolute pixel values.
- `modelPreprocessMovie`
- - Main processing method for calciumImagingAnalysis. Performs motion correction, spatial filtering, cropping, down-sampling, and relative fluorescence calculations. If using Inscopix nVista 1.0 or 2.0, also will correct for dropped frames.
+ - Main processing method for {{ site.name }}. Performs motion correction, spatial filtering, cropping, down-sampling, and relative fluorescence calculations. If using Inscopix nVista 1.0 or 2.0, also will correct for dropped frames.
- `modelModifyMovies`
- GUI that allows users to remove movie regions not relevant to cell extraction.
- `modelExtractSignalsFromMovie`
diff --git a/docs/make_docs.py b/docs/make_docs.py
index 8b15522..12fceab 100644
--- a/docs/make_docs.py
+++ b/docs/make_docs.py
@@ -19,6 +19,7 @@
# pip install mkdocs
# pip install mdx_truly_sane_lists
# pip install pymdown-extensions
+# pip install mkdocs-macros-plugin
# If you want to view the docs before building
# mkdocs serve
diff --git a/docs/mkdocs.yml b/docs/mkdocs.yml
index cd3b15f..113add5 100644
--- a/docs/mkdocs.yml
+++ b/docs/mkdocs.yml
@@ -1,31 +1,61 @@
-site_name: CIAtah (calciumImagingAnalysis [ciapkg])
+site_name: CIAtah
# For online, to end in /data instead of /data.html
use_directory_urls: true
theme:
name: readthedocs
nav_style: dark
- highlightjs: true
- hljs_style: github
- hljs_languages:
- - matlab
- - python
+ # highlightjs: true
+ highlightjs: false
+ # hljs_style: github
+ # hljs_languages:
+ # - matlab
+ # - python
# navigation_depth: 3
shortcuts:
help: 191 # ?
next: 78 # n
previous: 80 # p
search: 83 # s
-markdown_extensions:
+ features:
+ - navigation.sections
+ - navigation.expand
+plugins:
+ - macros
+markdown_extensions:
- toc:
permalink: true
+ # - codehilite
- pymdownx.magiclink
- mdx_truly_sane_lists
+ - pymdownx.highlight
+ - pymdownx.superfences
+ - pymdownx.inlinehilite
+ - pymdownx.highlight:
+ linenums: true
+ linenums_style: pymdownx-inline
+ use_pygments: true
+ css_class: 'codehilite'
+ guess_lang: false
# - markdown.extensions.toc:
# slugify: !!python/name:pymdownx.slugs.uslugify
# permalink: "\ue157"
# - codehilite
+extra_javascript:
+ # - https://cdnjs.cloudflare.com/ajax/libs/highlight.js/10.1.1/highlight.min.js
+ # - javascripts/config.js
extra_css:
+ # - https://cdnjs.cloudflare.com/ajax/libs/highlight.js/10.1.1/styles/default.min.css
- css/extra.css
+ - css/pygments.css
+
+# VARIABLES TO USE THROUGHOUT THE WEBSITE
+extra:
+ site:
+ name: CIAtah
+ namelow: ciatah
+ nameold: calciumImagingAnalysis
+
+# NAVIGATION
nav:
- Home: index.md
- One-page readme: alldocs.md
@@ -47,17 +77,24 @@ nav:
- ciapkg: api_ciapkg.md
- Help:
- Issues and fixes: help_issues.md
- - Analysis methods: help_analysis_methods.md
+ -
-******************************************
+
## Removing cells not within brain region with `modelModifyRegionAnalysis`
@@ -191,7 +226,7 @@ If the imaging field-of-view includes cells from other brain regions, they can b

-******************************************
+
## Cross-session cell alignment with `computeMatchObjBtwnTrials`
@@ -221,4 +256,4 @@ To evaluate how well cross-session alignment works, `computeMatchObjBtwnTrials`
### Save cross-session cell alignment with `modelSaveMatchObjBtwnTrials`
-Users can save out the alignment structure by running `modelSaveMatchObjBtwnTrials`. This will allow users to select a folder where `calciumImagingAnalysis` will save a MAT-file with the alignment structure information for each animal.
\ No newline at end of file
+Users can save out the alignment structure by running `modelSaveMatchObjBtwnTrials`. This will allow users to select a folder where `{{ site.name }}` will save a MAT-file with the alignment structure information for each animal.
\ No newline at end of file
diff --git a/docs/docs/pipeline_overview.md b/docs/docs/pipeline_overview.md
index c56cdbc..076ac0d 100644
--- a/docs/docs/pipeline_overview.md
+++ b/docs/docs/pipeline_overview.md
@@ -5,14 +5,14 @@ The general pipeline for processing calcium imaging data is below. This reposito

-To start using the `calciumImagingAnalysis` software package, enter the following into the MATLAB command window.
+To start using the `{{ site.name }}` software package, enter the following into the MATLAB command window.
```Matlab
% Loads all directories
loadBatchFxns;
% Loads the class into an object.
-obj = calciumImagingAnalysis;
+obj = ciatah;
% Open the class menu
obj % then hit enter, no semicolon!
@@ -23,14 +23,14 @@ obj.runPipeline; % then hit enter!
The general order of functions that users should run is ([optional] are those not critical for most datasets):
- `loadDependencies`
- - If user is running calciumImagingAnalysis for the first time, this module has several options to download and load CNMF/CNMF-E code for cell extraction, Fiji for viewing/modifying videos (using Miji), and test data from a miniature microscope experiment.
+ - If user is running {{ site.name }} for the first time, this module has several options to download and load CNMF/CNMF-E code for cell extraction, Fiji for viewing/modifying videos (using Miji), and test data from a miniature microscope experiment.
- `modelDownsampleRawMovies` [optional]
- If users have raw calcium imaging data that needs to be spatially downsampled, e.g. raw data from Inscopix nVista software.
- `modelAddNewFolders`
- Users should always use this method first, used to add folders to the current class object.
- - For example, if users ran `example_downloadTestData.m`, then add the folder `[githubRepoPath]\data\2014_04_01_p203_m19_check01_raw` where `githubRepoPath` is the absolute path to the current `calciumImagingAnalysis` repository.
+ - For example, if users ran `example_downloadTestData.m`, then add the folder `[githubRepoPath]\data\2014_04_01_p203_m19_check01_raw` where `githubRepoPath` is the absolute path to the current `{{ site.name }}` repository.
- `viewMovie`
- - Users should check that calciumImagingAnalysis loads their movies correctly and that Miji is working.
+ - Users should check that {{ site.name }} loads their movies correctly and that Miji is working.
- Users can view movies from disk, which allows checking of very large movies quickly.
- Remember to check that `Imaging movie regexp:` (regular expression class uses to find user movies within given folders) setting matches name of movies currently in repository.
- `viewMovieRegistrationTest` [optional]
@@ -38,7 +38,7 @@ The general order of functions that users should run is ([optional] are those no
- `tregRunX` folders (where `X` is a number) contain details of each run setting. Delete from analysis folder if don't need outputs later.
- Remember to adjust contrast in resulting montage movies since different filtering will change the absolute pixel values.
- `modelPreprocessMovie`
- - Main processing method for calciumImagingAnalysis. Performs motion correction, spatial filtering, cropping, down-sampling, and relative fluorescence calculations. If using Inscopix nVista 1.0 or 2.0, also will correct for dropped frames.
+ - Main processing method for {{ site.name }}. Performs motion correction, spatial filtering, cropping, down-sampling, and relative fluorescence calculations. If using Inscopix nVista 1.0 or 2.0, also will correct for dropped frames.
- `modelModifyMovies`
- GUI that allows users to remove movie regions not relevant to cell extraction.
- `modelExtractSignalsFromMovie`
diff --git a/docs/make_docs.py b/docs/make_docs.py
index 8b15522..12fceab 100644
--- a/docs/make_docs.py
+++ b/docs/make_docs.py
@@ -19,6 +19,7 @@
# pip install mkdocs
# pip install mdx_truly_sane_lists
# pip install pymdown-extensions
+# pip install mkdocs-macros-plugin
# If you want to view the docs before building
# mkdocs serve
diff --git a/docs/mkdocs.yml b/docs/mkdocs.yml
index cd3b15f..113add5 100644
--- a/docs/mkdocs.yml
+++ b/docs/mkdocs.yml
@@ -1,31 +1,61 @@
-site_name: CIAtah (calciumImagingAnalysis [ciapkg])
+site_name: CIAtah
# For online, to end in /data instead of /data.html
use_directory_urls: true
theme:
name: readthedocs
nav_style: dark
- highlightjs: true
- hljs_style: github
- hljs_languages:
- - matlab
- - python
+ # highlightjs: true
+ highlightjs: false
+ # hljs_style: github
+ # hljs_languages:
+ # - matlab
+ # - python
# navigation_depth: 3
shortcuts:
help: 191 # ?
next: 78 # n
previous: 80 # p
search: 83 # s
-markdown_extensions:
+ features:
+ - navigation.sections
+ - navigation.expand
+plugins:
+ - macros
+markdown_extensions:
- toc:
permalink: true
+ # - codehilite
- pymdownx.magiclink
- mdx_truly_sane_lists
+ - pymdownx.highlight
+ - pymdownx.superfences
+ - pymdownx.inlinehilite
+ - pymdownx.highlight:
+ linenums: true
+ linenums_style: pymdownx-inline
+ use_pygments: true
+ css_class: 'codehilite'
+ guess_lang: false
# - markdown.extensions.toc:
# slugify: !!python/name:pymdownx.slugs.uslugify
# permalink: "\ue157"
# - codehilite
+extra_javascript:
+ # - https://cdnjs.cloudflare.com/ajax/libs/highlight.js/10.1.1/highlight.min.js
+ # - javascripts/config.js
extra_css:
+ # - https://cdnjs.cloudflare.com/ajax/libs/highlight.js/10.1.1/styles/default.min.css
- css/extra.css
+ - css/pygments.css
+
+# VARIABLES TO USE THROUGHOUT THE WEBSITE
+extra:
+ site:
+ name: CIAtah
+ namelow: ciatah
+ nameold: calciumImagingAnalysis
+
+# NAVIGATION
nav:
- Home: index.md
- One-page readme: alldocs.md
@@ -47,17 +77,24 @@ nav:
- ciapkg: api_ciapkg.md
- Help:
- Issues and fixes: help_issues.md
- - Analysis methods: help_analysis_methods.md
+ - Data
: black
- Inscopix: help_inscopix.md
- - Large movie analysis: help_large_movie_analysis.md
+ - Neurodata Without Borders: help_nwb.md
+
+ - Interface
: blank
- Movie display and noise: help_contrast.md
- - Cross-session alignemnt: help_cross_session_alignment.md
- - Manual cell sorting: help_manual_cell_sorting.md
+
+ - Analysis
: blank
+ - Analysis methods: help_analysis_methods.md
+ - Large movie analysis: help_large_movie_analysis.md
+ - Stripe removal: help_stripe_removal.md
- Improving signal-to-noise: help_snr.md
- - Motion correction: help_motion_correction.md
- Spatial filtering: help_spatial_filtering.md
- - Stripe removal: help_stripe_removal.md
- Temporal downsampling: help_temporal_downsampling.md
+ - Motion correction: help_motion_correction.md
+ - Cell extraction: help_cell_extraction.md
+ - Manual cell sorting: help_manual_cell_sorting.md
+ - Cross-session alignemnt: help_cross_session_alignment.md
- Animal tracking: help_animal_tracking.md
- Misc:
- Acknowledgments: acknowledgments.md
diff --git a/docs/mkdocs_local.yml b/docs/mkdocs_local.yml
index b666e40..f05031d 100644
--- a/docs/mkdocs_local.yml
+++ b/docs/mkdocs_local.yml
@@ -20,6 +20,8 @@ markdown_extensions:
permalink: true
- pymdownx.magiclink
- mdx_truly_sane_lists
+ - pymdownx.highlight:
+ linenums: true
# - markdown.extensions.toc:
# slugify: !!python/name:pymdownx.slugs.uslugify
# permalink: "\ue157"
 +
+
+
+ +
+ -- This user guide contains instructions to setup, run, and troubleshoot `CIAtah`.
-- Note: `CIAtah` is a class (`ciatah` or `calciumImagingAnalysis` within MATLAB) with various GUIs to allow processing of calcium imaging data. In addition, users can access the underlying `CIAtah` functions to make custom workflows. See [Custom command-line pipelines](api_example_pipeline.md).
+- This user guide contains instructions to setup, run, and troubleshoot `{{ site.name }}`.
+- Note: `{{ site.name }}` is a class (`{{ site.namelow }}` or `calciumImagingAnalysis` within MATLAB) with various GUIs to allow processing of calcium imaging data. In addition, users can access the underlying `{{ site.name }}` functions to make custom workflows. See [Custom command-line pipelines](api_example_pipeline.md).
- Read my overview of calcium imaging analysis methods at [Calcium imaging cell identification and fluorescence activity trace reconstruction, part 1](https://bahanonu.com/brain/#c20181209).
-- This user guide contains instructions to setup, run, and troubleshoot `CIAtah`.
-- Note: `CIAtah` is a class (`ciatah` or `calciumImagingAnalysis` within MATLAB) with various GUIs to allow processing of calcium imaging data. In addition, users can access the underlying `CIAtah` functions to make custom workflows. See [Custom command-line pipelines](api_example_pipeline.md).
+- This user guide contains instructions to setup, run, and troubleshoot `{{ site.name }}`.
+- Note: `{{ site.name }}` is a class (`{{ site.namelow }}` or `calciumImagingAnalysis` within MATLAB) with various GUIs to allow processing of calcium imaging data. In addition, users can access the underlying `{{ site.name }}` functions to make custom workflows. See [Custom command-line pipelines](api_example_pipeline.md).
- Read my overview of calcium imaging analysis methods at [Calcium imaging cell identification and fluorescence activity trace reconstruction, part 1](https://bahanonu.com/brain/#c20181209).

 +
+
+
+ -******************************************
+
## Removing cells not within brain region with `modelModifyRegionAnalysis`
@@ -191,7 +226,7 @@ If the imaging field-of-view includes cells from other brain regions, they can b

-******************************************
+
## Cross-session cell alignment with `computeMatchObjBtwnTrials`
@@ -221,4 +256,4 @@ To evaluate how well cross-session alignment works, `computeMatchObjBtwnTrials`
### Save cross-session cell alignment with `modelSaveMatchObjBtwnTrials`
-Users can save out the alignment structure by running `modelSaveMatchObjBtwnTrials`. This will allow users to select a folder where `calciumImagingAnalysis` will save a MAT-file with the alignment structure information for each animal.
\ No newline at end of file
+Users can save out the alignment structure by running `modelSaveMatchObjBtwnTrials`. This will allow users to select a folder where `{{ site.name }}` will save a MAT-file with the alignment structure information for each animal.
\ No newline at end of file
diff --git a/docs/docs/pipeline_overview.md b/docs/docs/pipeline_overview.md
index c56cdbc..076ac0d 100644
--- a/docs/docs/pipeline_overview.md
+++ b/docs/docs/pipeline_overview.md
@@ -5,14 +5,14 @@ The general pipeline for processing calcium imaging data is below. This reposito

-To start using the `calciumImagingAnalysis` software package, enter the following into the MATLAB command window.
+To start using the `{{ site.name }}` software package, enter the following into the MATLAB command window.
```Matlab
% Loads all directories
loadBatchFxns;
% Loads the class into an object.
-obj = calciumImagingAnalysis;
+obj = ciatah;
% Open the class menu
obj % then hit enter, no semicolon!
@@ -23,14 +23,14 @@ obj.runPipeline; % then hit enter!
The general order of functions that users should run is ([optional] are those not critical for most datasets):
- `loadDependencies`
- - If user is running calciumImagingAnalysis for the first time, this module has several options to download and load CNMF/CNMF-E code for cell extraction, Fiji for viewing/modifying videos (using Miji), and test data from a miniature microscope experiment.
+ - If user is running {{ site.name }} for the first time, this module has several options to download and load CNMF/CNMF-E code for cell extraction, Fiji for viewing/modifying videos (using Miji), and test data from a miniature microscope experiment.
- `modelDownsampleRawMovies` [optional]
- If users have raw calcium imaging data that needs to be spatially downsampled, e.g. raw data from Inscopix nVista software.
- `modelAddNewFolders`
- Users should always use this method first, used to add folders to the current class object.
- - For example, if users ran `example_downloadTestData.m`, then add the folder `[githubRepoPath]\data\2014_04_01_p203_m19_check01_raw` where `githubRepoPath` is the absolute path to the current `calciumImagingAnalysis` repository.
+ - For example, if users ran `example_downloadTestData.m`, then add the folder `[githubRepoPath]\data\2014_04_01_p203_m19_check01_raw` where `githubRepoPath` is the absolute path to the current `{{ site.name }}` repository.
- `viewMovie`
- - Users should check that calciumImagingAnalysis loads their movies correctly and that Miji is working.
+ - Users should check that {{ site.name }} loads their movies correctly and that Miji is working.
- Users can view movies from disk, which allows checking of very large movies quickly.
- Remember to check that `Imaging movie regexp:` (regular expression class uses to find user movies within given folders) setting matches name of movies currently in repository.
- `viewMovieRegistrationTest` [optional]
@@ -38,7 +38,7 @@ The general order of functions that users should run is ([optional] are those no
- `tregRunX` folders (where `X` is a number) contain details of each run setting. Delete from analysis folder if don't need outputs later.
- Remember to adjust contrast in resulting montage movies since different filtering will change the absolute pixel values.
- `modelPreprocessMovie`
- - Main processing method for calciumImagingAnalysis. Performs motion correction, spatial filtering, cropping, down-sampling, and relative fluorescence calculations. If using Inscopix nVista 1.0 or 2.0, also will correct for dropped frames.
+ - Main processing method for {{ site.name }}. Performs motion correction, spatial filtering, cropping, down-sampling, and relative fluorescence calculations. If using Inscopix nVista 1.0 or 2.0, also will correct for dropped frames.
- `modelModifyMovies`
- GUI that allows users to remove movie regions not relevant to cell extraction.
- `modelExtractSignalsFromMovie`
diff --git a/docs/make_docs.py b/docs/make_docs.py
index 8b15522..12fceab 100644
--- a/docs/make_docs.py
+++ b/docs/make_docs.py
@@ -19,6 +19,7 @@
# pip install mkdocs
# pip install mdx_truly_sane_lists
# pip install pymdown-extensions
+# pip install mkdocs-macros-plugin
# If you want to view the docs before building
# mkdocs serve
diff --git a/docs/mkdocs.yml b/docs/mkdocs.yml
index cd3b15f..113add5 100644
--- a/docs/mkdocs.yml
+++ b/docs/mkdocs.yml
@@ -1,31 +1,61 @@
-site_name: CIAtah (calciumImagingAnalysis [ciapkg])
+site_name: CIAtah
# For online, to end in /data instead of /data.html
use_directory_urls: true
theme:
name: readthedocs
nav_style: dark
- highlightjs: true
- hljs_style: github
- hljs_languages:
- - matlab
- - python
+ # highlightjs: true
+ highlightjs: false
+ # hljs_style: github
+ # hljs_languages:
+ # - matlab
+ # - python
# navigation_depth: 3
shortcuts:
help: 191 # ?
next: 78 # n
previous: 80 # p
search: 83 # s
-markdown_extensions:
+ features:
+ - navigation.sections
+ - navigation.expand
+plugins:
+ - macros
+markdown_extensions:
- toc:
permalink: true
+ # - codehilite
- pymdownx.magiclink
- mdx_truly_sane_lists
+ - pymdownx.highlight
+ - pymdownx.superfences
+ - pymdownx.inlinehilite
+ - pymdownx.highlight:
+ linenums: true
+ linenums_style: pymdownx-inline
+ use_pygments: true
+ css_class: 'codehilite'
+ guess_lang: false
# - markdown.extensions.toc:
# slugify: !!python/name:pymdownx.slugs.uslugify
# permalink: "\ue157"
# - codehilite
+extra_javascript:
+ # - https://cdnjs.cloudflare.com/ajax/libs/highlight.js/10.1.1/highlight.min.js
+ # - javascripts/config.js
extra_css:
+ # - https://cdnjs.cloudflare.com/ajax/libs/highlight.js/10.1.1/styles/default.min.css
- css/extra.css
+ - css/pygments.css
+
+# VARIABLES TO USE THROUGHOUT THE WEBSITE
+extra:
+ site:
+ name: CIAtah
+ namelow: ciatah
+ nameold: calciumImagingAnalysis
+
+# NAVIGATION
nav:
- Home: index.md
- One-page readme: alldocs.md
@@ -47,17 +77,24 @@ nav:
- ciapkg: api_ciapkg.md
- Help:
- Issues and fixes: help_issues.md
- - Analysis methods: help_analysis_methods.md
+ -
-******************************************
+
## Removing cells not within brain region with `modelModifyRegionAnalysis`
@@ -191,7 +226,7 @@ If the imaging field-of-view includes cells from other brain regions, they can b

-******************************************
+
## Cross-session cell alignment with `computeMatchObjBtwnTrials`
@@ -221,4 +256,4 @@ To evaluate how well cross-session alignment works, `computeMatchObjBtwnTrials`
### Save cross-session cell alignment with `modelSaveMatchObjBtwnTrials`
-Users can save out the alignment structure by running `modelSaveMatchObjBtwnTrials`. This will allow users to select a folder where `calciumImagingAnalysis` will save a MAT-file with the alignment structure information for each animal.
\ No newline at end of file
+Users can save out the alignment structure by running `modelSaveMatchObjBtwnTrials`. This will allow users to select a folder where `{{ site.name }}` will save a MAT-file with the alignment structure information for each animal.
\ No newline at end of file
diff --git a/docs/docs/pipeline_overview.md b/docs/docs/pipeline_overview.md
index c56cdbc..076ac0d 100644
--- a/docs/docs/pipeline_overview.md
+++ b/docs/docs/pipeline_overview.md
@@ -5,14 +5,14 @@ The general pipeline for processing calcium imaging data is below. This reposito

-To start using the `calciumImagingAnalysis` software package, enter the following into the MATLAB command window.
+To start using the `{{ site.name }}` software package, enter the following into the MATLAB command window.
```Matlab
% Loads all directories
loadBatchFxns;
% Loads the class into an object.
-obj = calciumImagingAnalysis;
+obj = ciatah;
% Open the class menu
obj % then hit enter, no semicolon!
@@ -23,14 +23,14 @@ obj.runPipeline; % then hit enter!
The general order of functions that users should run is ([optional] are those not critical for most datasets):
- `loadDependencies`
- - If user is running calciumImagingAnalysis for the first time, this module has several options to download and load CNMF/CNMF-E code for cell extraction, Fiji for viewing/modifying videos (using Miji), and test data from a miniature microscope experiment.
+ - If user is running {{ site.name }} for the first time, this module has several options to download and load CNMF/CNMF-E code for cell extraction, Fiji for viewing/modifying videos (using Miji), and test data from a miniature microscope experiment.
- `modelDownsampleRawMovies` [optional]
- If users have raw calcium imaging data that needs to be spatially downsampled, e.g. raw data from Inscopix nVista software.
- `modelAddNewFolders`
- Users should always use this method first, used to add folders to the current class object.
- - For example, if users ran `example_downloadTestData.m`, then add the folder `[githubRepoPath]\data\2014_04_01_p203_m19_check01_raw` where `githubRepoPath` is the absolute path to the current `calciumImagingAnalysis` repository.
+ - For example, if users ran `example_downloadTestData.m`, then add the folder `[githubRepoPath]\data\2014_04_01_p203_m19_check01_raw` where `githubRepoPath` is the absolute path to the current `{{ site.name }}` repository.
- `viewMovie`
- - Users should check that calciumImagingAnalysis loads their movies correctly and that Miji is working.
+ - Users should check that {{ site.name }} loads their movies correctly and that Miji is working.
- Users can view movies from disk, which allows checking of very large movies quickly.
- Remember to check that `Imaging movie regexp:` (regular expression class uses to find user movies within given folders) setting matches name of movies currently in repository.
- `viewMovieRegistrationTest` [optional]
@@ -38,7 +38,7 @@ The general order of functions that users should run is ([optional] are those no
- `tregRunX` folders (where `X` is a number) contain details of each run setting. Delete from analysis folder if don't need outputs later.
- Remember to adjust contrast in resulting montage movies since different filtering will change the absolute pixel values.
- `modelPreprocessMovie`
- - Main processing method for calciumImagingAnalysis. Performs motion correction, spatial filtering, cropping, down-sampling, and relative fluorescence calculations. If using Inscopix nVista 1.0 or 2.0, also will correct for dropped frames.
+ - Main processing method for {{ site.name }}. Performs motion correction, spatial filtering, cropping, down-sampling, and relative fluorescence calculations. If using Inscopix nVista 1.0 or 2.0, also will correct for dropped frames.
- `modelModifyMovies`
- GUI that allows users to remove movie regions not relevant to cell extraction.
- `modelExtractSignalsFromMovie`
diff --git a/docs/make_docs.py b/docs/make_docs.py
index 8b15522..12fceab 100644
--- a/docs/make_docs.py
+++ b/docs/make_docs.py
@@ -19,6 +19,7 @@
# pip install mkdocs
# pip install mdx_truly_sane_lists
# pip install pymdown-extensions
+# pip install mkdocs-macros-plugin
# If you want to view the docs before building
# mkdocs serve
diff --git a/docs/mkdocs.yml b/docs/mkdocs.yml
index cd3b15f..113add5 100644
--- a/docs/mkdocs.yml
+++ b/docs/mkdocs.yml
@@ -1,31 +1,61 @@
-site_name: CIAtah (calciumImagingAnalysis [ciapkg])
+site_name: CIAtah
# For online, to end in /data instead of /data.html
use_directory_urls: true
theme:
name: readthedocs
nav_style: dark
- highlightjs: true
- hljs_style: github
- hljs_languages:
- - matlab
- - python
+ # highlightjs: true
+ highlightjs: false
+ # hljs_style: github
+ # hljs_languages:
+ # - matlab
+ # - python
# navigation_depth: 3
shortcuts:
help: 191 # ?
next: 78 # n
previous: 80 # p
search: 83 # s
-markdown_extensions:
+ features:
+ - navigation.sections
+ - navigation.expand
+plugins:
+ - macros
+markdown_extensions:
- toc:
permalink: true
+ # - codehilite
- pymdownx.magiclink
- mdx_truly_sane_lists
+ - pymdownx.highlight
+ - pymdownx.superfences
+ - pymdownx.inlinehilite
+ - pymdownx.highlight:
+ linenums: true
+ linenums_style: pymdownx-inline
+ use_pygments: true
+ css_class: 'codehilite'
+ guess_lang: false
# - markdown.extensions.toc:
# slugify: !!python/name:pymdownx.slugs.uslugify
# permalink: "\ue157"
# - codehilite
+extra_javascript:
+ # - https://cdnjs.cloudflare.com/ajax/libs/highlight.js/10.1.1/highlight.min.js
+ # - javascripts/config.js
extra_css:
+ # - https://cdnjs.cloudflare.com/ajax/libs/highlight.js/10.1.1/styles/default.min.css
- css/extra.css
+ - css/pygments.css
+
+# VARIABLES TO USE THROUGHOUT THE WEBSITE
+extra:
+ site:
+ name: CIAtah
+ namelow: ciatah
+ nameold: calciumImagingAnalysis
+
+# NAVIGATION
nav:
- Home: index.md
- One-page readme: alldocs.md
@@ -47,17 +77,24 @@ nav:
- ciapkg: api_ciapkg.md
- Help:
- Issues and fixes: help_issues.md
- - Analysis methods: help_analysis_methods.md
+ -