diff --git a/.github/workflows/docs.yml b/.github/workflows/docs.yml
new file mode 100644
index 00000000..4c59603a
--- /dev/null
+++ b/.github/workflows/docs.yml
@@ -0,0 +1,33 @@
+name: Documentation
+
+on:
+ push:
+ branches:
+ - 'master'
+ - 'release-'
+ tags: '*'
+ pull_request:
+
+jobs:
+ build:
+ runs-on: ubuntu-latest
+ steps:
+ - uses: actions/checkout@v2
+ - name: Cache artifacts
+ uses: actions/cache@v1
+ env:
+ cache-name: cache-artifacts
+ with:
+ path: ~/.julia/artifacts
+ key: ${{ runner.os }}-test-${{ env.cache-name }}-${{ hashFiles('**/Project.toml') }}
+ restore-keys: |
+ ${{ runner.os }}-test-${{ env.cache-name }}-
+ ${{ runner.os }}-test-
+ ${{ runner.os }}-
+ - name: Install dependencies
+ run: julia --project=docs/ -e 'using Pkg; Pkg.develop(PackageSpec(path=pwd())); Pkg.instantiate()'
+ - name: Build and deploy
+ env:
+ GITHUB_TOKEN: ${{ secrets.GITHUB_TOKEN }}
+ DOCUMENTER_KEY: ${{ secrets.DOCUMENTER_KEY }} # https://github.com/JuliaDocs/Documenter.jl/issues/1177
+ run: julia --project=docs/ docs/make.jl
diff --git a/docs/.gitignore b/docs/.gitignore
index 9c6edfbd..3f8b53fe 100644
--- a/docs/.gitignore
+++ b/docs/.gitignore
@@ -1,3 +1,4 @@
build/
site/
-src/generated/*
+src/democards
+/Manifest.toml
diff --git a/docs/Project.toml b/docs/Project.toml
new file mode 100644
index 00000000..d7317d0c
--- /dev/null
+++ b/docs/Project.toml
@@ -0,0 +1,18 @@
+[deps]
+Augmentor = "02898b10-1f73-11ea-317c-6393d7073e15"
+DemoCards = "311a05b2-6137-4a5a-b473-18580a3d38b5"
+Documenter = "e30172f5-a6a5-5a46-863b-614d45cd2de4"
+FileIO = "5789e2e9-d7fb-5bc7-8068-2c6fae9b9549"
+ImageCore = "a09fc81d-aa75-5fe9-8630-4744c3626534"
+ImageDraw = "4381153b-2b60-58ae-a1ba-fd683676385f"
+ImageMagick = "6218d12a-5da1-5696-b52f-db25d2ecc6d1"
+ImageShow = "4e3cecfd-b093-5904-9786-8bbb286a6a31"
+Images = "916415d5-f1e6-5110-898d-aaa5f9f070e0"
+MLDatasets = "eb30cadb-4394-5ae3-aed4-317e484a6458"
+MosaicViews = "e94cdb99-869f-56ef-bcf0-1ae2bcbe0389"
+OffsetArrays = "6fe1bfb0-de20-5000-8ca7-80f57d26f881"
+PaddedViews = "5432bcbf-9aad-5242-b902-cca2824c8663"
+
+[compat]
+DemoCards = "0.2"
+Documenter = "0.24"
diff --git a/docs/examples/examples/assets/mnist_elastic.gif b/docs/examples/examples/assets/mnist_elastic.gif
new file mode 100644
index 00000000..e119f20f
Binary files /dev/null and b/docs/examples/examples/assets/mnist_elastic.gif differ
diff --git a/docs/examples/examples/mnist_elastic.md b/docs/examples/examples/mnist_elastic.md
new file mode 100644
index 00000000..53b70c25
--- /dev/null
+++ b/docs/examples/examples/mnist_elastic.md
@@ -0,0 +1,125 @@
+---
+title: Elastic distortion to MNIST images
+id: mnist_elastic
+cover: assets/mnist_elastic.gif
+---
+
+
+In this example we are going to use Augmentor on the famous **MNIST database of handwritten
+digits** [^MNIST1998] to reproduce the elastic distortions discussed in [^SIMARD2003].
+
+It may be interesting to point out, that the way Augmentor implements distortions is a little
+different to how it is described by the authors of the paper. This is for a couple of reasons,
+most notably that we want the parameters for our deformations to be independent of the size of
+image it is applied on. As a consequence the parameter-numbers specified in the paper are not
+1-to-1 transferable to Augmentor.
+
+If the effects are sensible for the dataset, then applying elastic distortions can be a really
+effective way to improve the generalization ability of the network. That said, our implementation
+of [`ElasticDistortion`](@ref) has a lot of possible parameters to choose from. To that end, we
+will introduce a simple strategy for interactively exploring the parameter space on our dataset of
+interest.
+
+## Loading the MNIST Trainingset
+
+In order to access and visualize the MNIST images we employ the help of two additional Julia
+packages. In the interest of time and space we will not go into great detail about their
+functionality. Feel free to click on their respective names to find out more information about the
+utility they can provide.
+
+- [Images.jl](https://github.com/JuliaImages/Images.jl) will provide us with the necessary tools
+ for working with image data in Julia.
+
+- [MLDatasets.jl](https://github.com/JuliaML/MLDatasets.jl) has an MNIST submodule that offers a
+ convenience interface to read the MNIST database.
+
+The function `MNIST.traintensor` returns the MNIST training images corresponding to the given
+indices as a multi-dimensional array. These images are stored in the native horizontal-major
+memory layout as a single floating point array, where all values are scaled to be between 0.0 and
+1.0.
+
+```@example mnist_elastic
+using Images, MLDatasets
+train_tensor = MNIST.traintensor()
+
+summary(train_tensor)
+```
+
+This horizontal-major format is the standard way of utilizing this dataset for training machine
+learning models. In this tutorial, however, we are more interested in working with the MNIST
+images as actual Julia images in vertical-major layout, and as black digits on white background.
+
+We can convert the "tensor" to a `Colorant` array using the provided function
+`MNIST.convert2image`. This way, Julia knows we are dealing with image data and can tell
+programming environments such as Juypter how to visualize it. If you are working in the terminal
+you may want to use the package
+[ImageInTerminal.jl](https://github.com/JuliaImages/ImageInTerminal.jl)
+
+```@example mnist_elastic
+train_images = MNIST.convert2image(train_tensor)
+
+train_images[:,:,1] # show first image
+```
+
+## Visualizing the Effects
+
+Before applying an operation (or pipeline of operations) on some dataset to train a network, we
+strongly recommend investing some time in selecting a decent set of hyper parameters for the
+operation(s). A useful tool for tasks like this is the package
+[Interact.jl](https://github.com/JuliaGizmos/Interact.jl). We will use this package to define a
+number of widgets for controlling the parameters to our operation.
+
+Note that while the code below only focuses on configuring the parameters of a single operation,
+specifically [`ElasticDistortion`](@ref), it could also be adapted to tweak a whole pipeline. Take
+a look at the corresponding section in [High-level Interface](@ref pipeline) for more information
+on how to define and use a pipeline.
+
+These two package will provide us with the capabilities to perform interactive visualisations in a
+jupyter notebook
+using Augmentor, Interact, Reactive
+
+The manipulate macro will turn the parameters of the
+loop into interactive widgets.
+
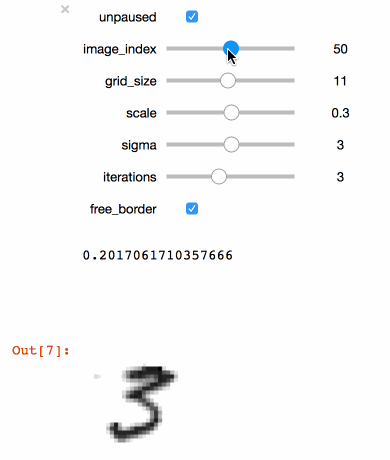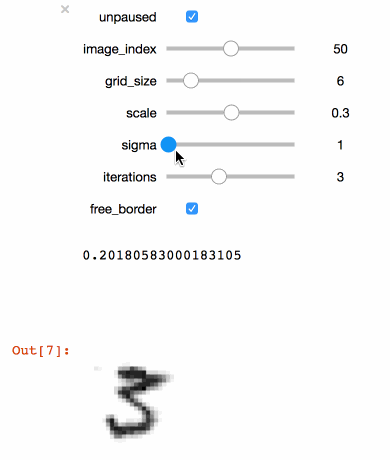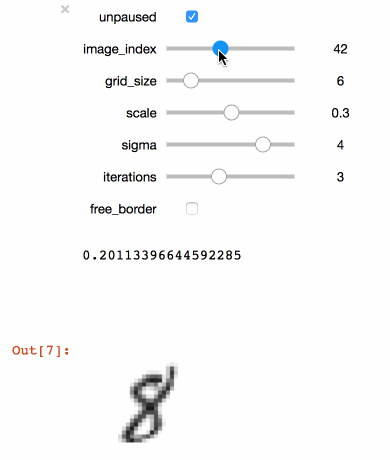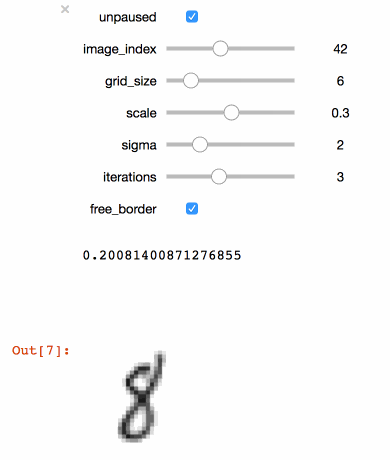
+```julia
+@manipulate for
+ unpaused = true,
+ ticks = fpswhen(signal(unpaused), 5.),
+ image_index = 1:100,
+ grid_size = 3:20,
+ scale = .1:.1:.5,
+ sigma = 1:5,
+ iterations = 1:6,
+ free_border = true op = ElasticDistortion(grid_size, grid_size, # equal width & height
+ sigma = sigma,
+ scale = scale,
+ iter = iterations,
+ border = free_border)
+ augment(train_images[:, :, image_index], op)
+end
+```
+
+Executing the code above in a Juypter notebook will result
+in the following interactive visualisation. You can now
+use the sliders to investigate the effects that different
+parameters have on the MNIST training images.
+
+!!! tip
+ You should always use your **training** set to do this
+ kind of visualisation (not the test test!). Otherwise
+ you are likely to achieve overly optimistic (i.e. biased)
+ results during training.
+
+
+
+Congratulations! With just a few simple lines of code, you
+created a simple interactive tool to visualize your image
+augmentation pipeline. Once you found a set of parameters that
+you think are appropriate for your dataset you can go ahead
+and train your model.
+
+## References
+
+[^MNIST1998]: LeCun, Yan, Corinna Cortes, Christopher J.C. Burges. ["The MNIST database of handwritten digits"](http://yann.lecun.com/exdb/mnist/) Website. 1998.
+
+[^SIMARD2003]: Simard, Patrice Y., David Steinkraus, and John C. Platt. ["Best practices for convolutional neural networks applied to visual document analysis."](https://www.microsoft.com/en-us/research/publication/best-practices-for-convolutional-neural-networks-applied-to-visual-document-analysis/) ICDAR. Vol. 3. 2003.
diff --git a/docs/examples/index.md b/docs/examples/index.md
new file mode 100644
index 00000000..70ea7bee
--- /dev/null
+++ b/docs/examples/index.md
@@ -0,0 +1,12 @@
+# [Tutorials](@id tutorials)
+
+Here we provide several tutorials that you may want to follow up and see how Augmentor is used in
+practice.
+
+{{{democards}}}
+
+## References
+
+[^MNIST1998]: LeCun, Yan, Corinna Cortes, Christopher J.C. Burges. ["The MNIST database of handwritten digits"](http://yann.lecun.com/exdb/mnist/) Website. 1998.
+
+[^SIMARD2003]: Simard, Patrice Y., David Steinkraus, and John C. Platt. ["Best practices for convolutional neural networks applied to visual document analysis."](https://www.microsoft.com/en-us/research/publication/best-practices-for-convolutional-neural-networks-applied-to-visual-document-analysis/) ICDAR. Vol. 3. 2003.
diff --git a/docs/exampleweaver.jl b/docs/exampleweaver.jl
deleted file mode 100644
index 8457a6a6..00000000
--- a/docs/exampleweaver.jl
+++ /dev/null
@@ -1,147 +0,0 @@
-"""
- module ExampleWeaver
-
-Uses the package `Weave` to generate `Documenter`-compatible
-markdown files, as well as pre-executed Juypter notebooks, from
-normal `.jl` scripts contained in the "examples/" subfolder of
-the package.
-
-The resulting markdown and notebook documents will be stored at
-"docs/src/generated/" of the package. Thus it is advised to add
-that folder to your **toplevel** `.gitignore` file. Do not put a
-`.gitignore` file into "docs/src/generated" itself, as that would
-affect the build documentation as well.
-
-Note the following additions to the usual `Weave`-compatible
-comment syntax that is supported for the ".jl" scripts in the
-"examples/" folder:
-
-- Lines that begin with `# md` will only be included
- in the markdown file (with the `# md` prefix removed)
-
-- Lines that begin with `# jp` will only be included
- in the Jupyter notebook (with the `# jp` prefix removed)
-"""
-module ExampleWeaver
-using Weave
-
-export
-
- listexamples,
- listmarkdown,
- listnotebooks,
-
- weave_markdown,
- weave_notebook,
- weave
-
-# --------------------------------------------------------------------
-
-const EXAMPLES_DIR = abspath(joinpath(@__DIR__, "..", "examples"))
-const GENERATED_DIR = abspath(joinpath(@__DIR__, "src", "generated"))
-
-function _listfiles(dir, ext, fullpath=false)
- fnames = filter(fname->splitext(fname)[2]==ext, readdir(dir))
- fullpath ? map(fname->joinpath(dir, fname), fnames) : fnames
-end
-
-listexamples(fullpath=false) = _listfiles(EXAMPLES_DIR, ".jl", fullpath)
-listmarkdown(fullpath=false) = _listfiles(GENERATED_DIR, ".md", fullpath)
-listnotebooks(fullpath=false) = _listfiles(GENERATED_DIR, ".ipynb", fullpath)
-
-# --------------------------------------------------------------------
-
-function weave_markdown(scriptname; overwrite=false)
- splitext(scriptname)[2] == ".jl" || return
- name = splitext(scriptname)[1]
- # define all required paths
- scriptpath = joinpath(EXAMPLES_DIR, scriptname)
- processed_scriptpath = joinpath(GENERATED_DIR, name * ".jl")
- jmdpath = joinpath(GENERATED_DIR, name * ".jmd")
- mdpath = joinpath(GENERATED_DIR, name * ".md")
- # if markdown file already exists, only overwrite if requested
- if isfile(mdpath) && !overwrite
- info("skipping markdown generation for \"$scriptname\" (file already exists)")
- return mdpath
- else
- info("generating markdown \"$(name*".md")\" for \"$scriptname\"")
- mkpath(GENERATED_DIR)
- end
- # load and pre-process script for markdown generation this
- # removes `# jp` and `#jp-only` lines and the `# md` prefix
- str_jl = readstring(scriptpath)
- str_jl = replace(str_jl, r"\n(#jp ).*", "")
- str_jl = replace(str_jl, r"\n.*(#jl-only)", "")
- str_jl = replace(str_jl, "\n#md ", "\n")
- write(processed_scriptpath, str_jl)
- # weave the .jl file into a .jmd file
- convert_doc(processed_scriptpath, jmdpath)
- # posprocess the .jmd and save it as .md for documenter
- str_md = readstring(jmdpath)
- str_md = replace(str_md, "```julia", "```@example $name")
- write(mdpath, str_md)
- # cleanup temporary files
- rm(processed_scriptpath)
- rm(jmdpath)
- # return path to final .md file
- mdpath
-end
-
-function weave_notebook(scriptname; overwrite=false, execute=true)
- splitext(scriptname)[2] == ".jl" || return
- name = splitext(scriptname)[1]
- # define all required paths
- scriptpath = joinpath(EXAMPLES_DIR, scriptname)
- processed_scriptpath = joinpath(GENERATED_DIR, name * ".jl")
- jppath = joinpath(GENERATED_DIR, name * ".ipynb")
- # if notebook file already exists, only overwrite if requested
- if isfile(jppath) && !overwrite
- info("skipping notebook generation for \"$scriptname\" (file already exists)")
- return jppath
- else
- info("generating notebook \"$(name*".ipynb")\" for \"$scriptname\"")
- mkpath(GENERATED_DIR)
- end
- # load and pre-process script for notebook generation this
- # removes `# md` and `#jp-only` lines and the `# jp` prefix
- str_jl = readstring(scriptpath)
- str_jl = replace(str_jl, r"\n(#md ).*", "")
- str_jl = replace(str_jl, r"\n.*(#jl-only)", "")
- str_jl = replace(str_jl, "\n#jp ", "\n")
- # additionally we slightly tweak the look of the references
- str_jl = replace(str_jl, r"\[\^(.*)\]:", s"**\1**:") # references
- str_jl = replace(str_jl, r"\[\^(.*)\]", s"[\1]") # citations
- write(processed_scriptpath, str_jl)
- # weave the .jl file into a .ipynb file
- convert_doc(processed_scriptpath, jppath)
- # execute notebook
- if execute
- sleep(1)
- info("executing and overwrite notebook \"$(name*".ipynb")\"")
- run(`jupyter-nbconvert --ExecutePreprocessor.timeout=-1 --to notebook --execute $(abspath(jppath)) --output $(name * ".ipynb")`)
- end
- # cleanup temporary files
- rm(processed_scriptpath)
- # return path to final .md file
- jppath
-end
-
-# --------------------------------------------------------------------
-
-function weave(scriptname; overwrite=false, execute=true)
- md = weave_markdown(scriptname; overwrite=overwrite)
- jp = weave_notebook(scriptname; overwrite=overwrite, execute=execute)
- md, jp
-end
-
-function weave(; kw...)
- mds = String[]; jps = String[]
- for scriptname in listexamples()
- md, jp = weave(scriptname; kw...)
- push!(mds, md)
- push!(jps, jp)
- end
- mds, jps
-end
-
-end # module
diff --git a/docs/make.jl b/docs/make.jl
index 6bc70b53..8ef36dc9 100644
--- a/docs/make.jl
+++ b/docs/make.jl
@@ -1,109 +1,57 @@
-using Documenter, Augmentor
+using Documenter, DemoCards
+using Augmentor
+using Random
+using MLDatasets
-# Autogenerate documentation markdown and jupyter notebooks
-# for all the scripts in the "examples/" subfolder.
-include("exampleweaver.jl")
-ExampleWeaver.weave(overwrite=false, execute=true)
+try
+ using ISICArchive
+catch
+ using Pkg
+ Pkg.add(url="https://github.com/Evizero/ISICArchive.jl.git", rev="master")
+ using ISICArchive
+end
+
+ENV["DATADEPS_ALWAYS_ACCEPT"] = true # MLDatasets
-# Define the documentation order of the operations. The whole
-# purpose of this vector is literally just to dictate in what
-# chronological order the operations are documented.
-op_fnames = [
- "flipx",
- "flipy",
- "rotate90",
- "rotate270",
- "rotate180",
- "rotate",
- "shearx",
- "sheary",
- "scale",
- "zoom",
- "elasticdistortion",
- "crop",
- "cropnative",
- "cropsize",
- "cropratio",
- "rcropratio",
- "resize",
- "converteltype",
- "mapfun",
- "aggmapfun",
- "splitchannels",
- "combinechannels",
- "permutedims",
- "reshape",
- "noop",
- "cacheimage",
- "either",
-]
-dict_order = Dict(fname * ".md" => i for (i, fname) in enumerate(op_fnames))
-myless(a, b) = dict_order[a] < dict_order[b]
+op_templates, op_theme = cardtheme("grid")
+operations, operations_cb = makedemos("operations", op_templates)
+examples_templates, examples_theme = cardtheme("list")
+examples, examples_cb = makedemos("examples", examples_templates)
-# --------------------------------------------------------------------
+format = Documenter.HTML(edit_link = "master",
+ prettyurls = get(ENV, "CI", nothing) == "true",
+ assets = [
+ joinpath("assets", "favicon.ico"),
+ joinpath("assets", "style.css"),
+ op_theme,
+ examples_theme
+ ]
+)
-srand(1337)
makedocs(
modules = [Augmentor],
- clean = false,
- format = :html,
- assets = [
- joinpath("assets", "favicon.ico"),
- joinpath("assets", "style.css"),
- ],
sitename = "Augmentor.jl",
authors = "Christof Stocker",
- linkcheck = !("skiplinks" in ARGS),
- pages = Any[
+ # linkcheck = true,
+ format = format,
+ pages = [
"Home" => "index.md",
"gettingstarted.md",
- "Introduction and Motivation" => Any[
+ "Introduction and Motivation" => [
"background.md",
"images.md",
],
- "User's Guide" => Any[
+ "User's Guide" => [
"interface.md",
- hide("operations.md", Any[joinpath("operations", fname) for fname in sort(readdir(joinpath(@__DIR__, "src", "operations")), lt = myless) if splitext(fname)[2] == ".md"]),
+ operations,
],
- "Tutorials" => joinpath.("generated", ExampleWeaver.listmarkdown()),
+ "Tutorials" => examples,
hide("Indices" => "indices.md"),
"LICENSE.md",
- ],
- html_prettyurls = !("local" in ARGS),
+ ]
)
-deploydocs(
- repo = "github.com/Evizero/Augmentor.jl.git",
- target = "build",
- julia = "0.6",
- deps = nothing,
- make = nothing,
-)
-
-# --------------------------------------------------------------------
-# Post-process the generated HTML files of the examples/tutorials
-# 1. Redirect "Edit on Github" link to the "examples/*.jl" file
-# 2. Add a link in the top right corner to the Juypter notebook
+operations_cb()
+examples_cb()
-build_dir = abspath(joinpath(@__DIR__, "build"))
-for markdownname in ExampleWeaver.listmarkdown()
- name = splitext(markdownname)[1]
- htmlpath = joinpath(build_dir, "generated", name, "index.html")
- str_html = readstring(htmlpath)
- # replace github url to .jl file
- str_html = replace(
- str_html,
- r"docs/src/generated/([^.]*)\.md",
- s"examples/\1.jl"
- )
- # insert link to jupyter notebook
- str_html = replace(
- str_html,
- r"()",
- s"\1 Juypter Notebook"
- )
- href = "https://nbviewer.jupyter.org/github/Evizero/Augmentor.jl/blob/gh-pages/generated/$(name * ".ipynb")"
- str_html = replace(str_html, "___HREFPLACEHOLDER___", href)
- # overwrite html file
- write(htmlpath, str_html)
-end
+deploydocs(repo = "github.com/Evizero/Augmentor.jl.git")
diff --git a/docs/operations/affine/config.json b/docs/operations/affine/config.json
new file mode 100644
index 00000000..96eead3e
--- /dev/null
+++ b/docs/operations/affine/config.json
@@ -0,0 +1,12 @@
+{
+ "title": "Affine Transformations",
+ "order": [
+ "flip.jl",
+ "rotate.jl",
+ "shear.jl",
+ "scale.jl",
+ "resize.jl",
+ "zoom.jl"
+ ],
+ "description": "A sizeable amount of the provided operations fall under the category of **affine transformations**. As such, they can be described using what is known as an [affine map](https://en.wikipedia.org/wiki/Affine_transformation), which are inherently compose-able if chained together. However, utilizing such a affine formulation requires (costly) interpolation, which may not always be needed to achieve the desired effect. For that reason do some of the operations below also provide a special purpose implementation to produce their specified result. Those are usually preferred over the affine formulation if sensible considering the complete pipeline."
+}
diff --git a/docs/operations/affine/flip.jl b/docs/operations/affine/flip.jl
new file mode 100644
index 00000000..4683ae3c
--- /dev/null
+++ b/docs/operations/affine/flip.jl
@@ -0,0 +1,37 @@
+# ---
+# title: Flip
+# cover: flip.gif
+# description: flip the input image horizontally or vertically
+# ---
+
+# [`FlipX`](@ref)/[`FlipY`](@ref) can be used to flip the input image horizontally/vertically.
+
+using Augmentor
+using ImageShow, ImageCore
+
+img_in = testpattern(RGB, ratio=0.5)
+
+mosaicview(
+ img_in,
+ augment(img_in, FlipX()),
+ augment(img_in, FlipY());
+ fillvalue=colorant"white", nrow=1, npad=10
+)
+
+# To perform a random flip, you can also pass the probablity to the constructor. For example, `FlipX(0.5)`
+# flips the image with half chance.
+
+# ## References
+
+#md # ```@docs
+#md # FlipX
+#md # FlipY
+#md # ```
+
+
+## save covers #src
+using ImageMagick #src
+using FileIO #src
+include(joinpath("..", "assets", "utilities.jl")) #src
+cover = make_gif(testpattern(RGB, ratio=0.5), Either(FlipX(), FlipY()), 4) #src
+ImageMagick.save("flip.gif", cover; fps=1) #src
diff --git a/docs/operations/affine/resize.jl b/docs/operations/affine/resize.jl
new file mode 100644
index 00000000..5bab5243
--- /dev/null
+++ b/docs/operations/affine/resize.jl
@@ -0,0 +1,30 @@
+# ---
+# title: Resize
+# cover: resize.gif
+# ---
+
+# Set the static size of the image
+
+using Augmentor
+using ImageShow, ImageCore
+
+img_in = testpattern(RGB, ratio=0.5)
+
+mosaicview(
+ img_in,
+ augment(img_in, Resize(240, 320));
+ fillvalue=colorant"white", nrow=1, npad=10
+)
+
+
+# ## References
+
+#md # ```@docs
+#md # Resize
+#md # ```
+
+## save covers #src
+using ImageMagick #src
+include(joinpath("..", "assets", "utilities.jl")) #src
+cover = make_gif(testpattern(RGB, ratio=0.5), Resize(240, 320), 2) #src
+ImageMagick.save("resize.gif", cover; fps=1) #src
diff --git a/docs/operations/affine/rotate.jl b/docs/operations/affine/rotate.jl
new file mode 100644
index 00000000..e6860e8c
--- /dev/null
+++ b/docs/operations/affine/rotate.jl
@@ -0,0 +1,54 @@
+# ---
+# title: Rotate
+# cover: rotate.gif
+# description: rotate image anticlockwise
+# ---
+
+# The type [`Rotate`](@ref) defines a generic anticlockwise rotation operation around the center
+# of the image. It is also possible to pass some abstract vector to the constructor, in which case
+# Augmentor will randomly sample one of its elements every time the operation is applied.
+
+using Augmentor
+using ImageShow, ImageCore
+using Random
+Random.seed!(0)
+
+img_in = testpattern(RGB, ratio=0.5)
+mosaicview(
+ img_in,
+
+ ## deterministic rotation
+ augment(img_in, Rotate(45)),
+
+ ## random rotation
+ augment(img_in, Rotate(-45:45));
+ fillvalue=colorant"white", nrow=1, npad=10
+)
+
+# Note that the output image size will be changed after rotation, [`CropNative`](@ref) can be particalually
+# useful to preserve the image size.
+
+mosaicview(
+ augment(img_in, Rotate(45)),
+ augment(img_in, Rotate(45) |> CropNative(axes(img_in)));
+ nrow=1, npad=10
+)
+
+# Rotation by some special degree (e.g.,90, 180 and 270) can be handled more efficiently without interpolation.
+# Compared to `Rotate(90)`, it is recommended to use [`Rotate90`](@ref) when possible. [`Rotate180`](@ref) and
+# [`Rotate270`](@ref) are available, too.
+
+# ## References
+
+#md # ```@docs
+#md # Rotate
+#md # Rotate90
+#md # Rotate180
+#md # Rotate270
+#md # ```
+
+## save covers #src
+using ImageMagick #src
+include(joinpath("..", "assets", "utilities.jl")) #src
+cover = make_gif(testpattern(RGB, ratio=0.5), Rotate(-20:20), 5) #src
+ImageMagick.save("rotate.gif", cover; fps=1) #src
diff --git a/docs/operations/affine/scale.jl b/docs/operations/affine/scale.jl
new file mode 100644
index 00000000..82954ed3
--- /dev/null
+++ b/docs/operations/affine/scale.jl
@@ -0,0 +1,44 @@
+# ---
+# title: Scale
+# cover: scale.gif
+# ---
+
+# Relatively resizing image
+
+using Augmentor
+using ImageShow, ImageCore
+using Random
+
+# In the case that only a single scale factor is specified, the
+# operation will assume that the intention is to scale all
+# dimensions uniformly by that factor.
+
+img_in = testpattern(RGB, ratio=0.5)
+
+mosaicview(
+ img_in,
+ augment(img_in, Scale(0.8)),
+ augment(img_in, Scale(0.8, 1));
+
+ fillvalue=colorant"white", nrow=1, npad=10
+)
+
+# It is also possible to pass some abstract vector(s) to the
+# constructor, in which case Augmentor will randomly sample one of
+# its elements every time the operation is applied.
+
+Random.seed!(1337)
+img_out = [augment(img_in, Scale(0.9:0.05:1.2)) for _ in 1:4]
+mosaicview(img_out...; fillvalue=colorant"white", nrow=2)
+
+# ## References
+
+#md # ```@docs
+#md # Scale
+#md # ```
+
+## save covers #src
+using ImageMagick #src
+include(joinpath("..", "assets", "utilities.jl")) #src
+cover = make_gif(testpattern(RGB, ratio=0.5), Scale(0.9:0.1:1.5), 5) #src
+ImageMagick.save("scale.gif", cover; fps=1) #src
diff --git a/docs/operations/affine/shear.jl b/docs/operations/affine/shear.jl
new file mode 100644
index 00000000..d705975f
--- /dev/null
+++ b/docs/operations/affine/shear.jl
@@ -0,0 +1,49 @@
+# ---
+# title: Shear
+# cover: shear.gif
+# description: shear the input image horizontally or vertically
+# ---
+
+# [`ShearX`](@ref)/[`ShearY`](@ref) can be used to shear the input image horizontally/vertically.
+# The input to the constructor can be a scalar or a vector. In the case of a vector, the transformation
+# will be a stochastic process.
+
+using Augmentor
+using ImageShow, ImageCore
+using Random
+Random.seed!(0)
+img_in = testpattern(RGB, ratio=0.5)
+
+mosaicview(
+ ## deterministic transformation
+ augment(img_in, ShearX(20)),
+ augment(img_in, ShearY(20)),
+
+ ## random transformation
+ augment(img_in, ShearX(-20:20)),
+ augment(img_in, ShearY(-20:20));
+
+ fillvalue=colorant"white", nrow=2, npad=10
+)
+
+# Note that the output image size will be changed after transformation, [`CropNative`](@ref) can be
+# particalually useful to preserve the image size.
+
+mosaicview(
+ augment(img_in, ShearX(10)),
+ augment(img_in, ShearX(10) |> CropNative(axes(img_in)));
+ fillvalue=colorant"white", nrow=1, npad=10
+)
+
+# ## References
+
+#md # ```@docs
+#md # ShearX
+#md # ShearY
+#md # ```
+
+## save covers #src
+using ImageMagick #src
+include(joinpath("..", "assets", "utilities.jl")) #src
+cover = make_gif(testpattern(RGB, ratio=0.5), Either(ShearX(-10:10), ShearY(-10:10)), 5) #src
+ImageMagick.save("shear.gif", cover; fps=1) #src
diff --git a/docs/operations/affine/zoom.jl b/docs/operations/affine/zoom.jl
new file mode 100644
index 00000000..48eb3d2f
--- /dev/null
+++ b/docs/operations/affine/zoom.jl
@@ -0,0 +1,44 @@
+# ---
+# title: Zoom
+# cover: zoom.gif
+# ---
+
+# Scale without resize
+
+using Augmentor
+using ImageShow, ImageCore
+using Random
+
+# In the case that only a single Zoom factor is specified, the
+# operation will assume that the intention is to Zoom all
+# dimensions uniformly by that factor.
+
+img_in = testpattern(RGB, ratio=0.5)
+
+mosaicview(
+ img_in,
+ augment(img_in, Zoom(1.3)),
+ augment(img_in, Zoom(1.3, 1));
+ fillvalue=colorant"white", nrow=1, npad=10
+)
+
+# It is also possible to pass some abstract vector(s) to the
+# constructor, in which case Augmentor will randomly sample one of
+# its elements every time the operation is applied.
+
+Random.seed!(1337)
+img_out = [augment(img_in, Zoom(0.9:0.05:1.2)) for _ in 1:4]
+
+mosaicview(img_out...; nrow=2)
+
+# ## References
+
+#md # ```@docs
+#md # Zoom
+#md # ```
+
+## save covers #src
+using ImageMagick #src
+include(joinpath("..", "assets", "utilities.jl")) #src
+cover = make_gif(testpattern(RGB, ratio=0.5), Zoom(0.9:0.1:1.5), 5) #src
+ImageMagick.save("zoom.gif", cover; fps=1) #src
diff --git a/docs/operations/assets/utilities.jl b/docs/operations/assets/utilities.jl
new file mode 100644
index 00000000..d47ea4d3
--- /dev/null
+++ b/docs/operations/assets/utilities.jl
@@ -0,0 +1,90 @@
+using Augmentor
+using ImageCore
+using Random
+using ImageCore: GenericImage
+
+"""
+ make_gif(img, pl, num_sample; random_seed=1337, kwargs...)
+
+Augment `img` with pipeline `pl` for num_sample times, and concatenate them into a 3 dimensional
+image.
+
+# Examples
+
+The main purpose of this function is to generate a 3-dimensional image so that we could save a gif
+cover using `ImageMagick.save`.
+
+```julia
+using Augmentor, ImageMagick
+cover = make_gif(testpattern(RGB), FlipX(), 2)
+ImageMagick.save("flipx.gif", cover; fps=1)
+```
+
+`img` can be a list of images, too. In this case, additional `kwargs` are passed to `mosaicview` so
+that you could control how images are ordered.
+
+```julia
+pl = ElasticDistortion(6, scale=0.3, border=true) |>
+ Rotate([10, -5, -3, 0, 3, 5, 10]) |>
+ ShearX(-10:10) * ShearY(-10:10) |>
+ CropSize(28, 28) |>
+ Zoom(0.9:0.1:1.2)
+
+n_samples, n_frames = 24, 10
+imgs = [MNIST.convert2image(MNIST.traintensor(i)) for i in 1:n_samples]
+preview = make_gif(imgs, pl, n_frames; nrow=1)
+```
+"""
+function make_gif(img::GenericImage, pl, num_sample; post_op=center_pad ∘ drawborder, random_seed=1337)
+ Random.seed!(random_seed)
+
+ fillvalue = oneunit(eltype(img))
+ frames = sym_paddedviews(
+ fillvalue,
+ post_op(img),
+ [post_op(augment(img, pl)) for _ in 1:num_sample-1]...
+ )
+ cat(frames..., dims=3)
+end
+
+function make_gif(img, pl, num_sample; post_op=drawborder, random_seed=1337, kwargs...)
+ fillvalue = oneunit(eltype(img[1]))
+
+ init_frame = mosaicview(post_op.(img); kwargs...)
+ frames = map(1:num_sample-1) do _
+ mosaicview(map(x->post_op(augment(x, pl)), img)...; kwargs...)
+ end
+
+ frames = sym_paddedviews(fillvalue, init_frame, frames...)
+ cat(frames..., dims=3)
+end
+
+"""
+ center_pad(img, sz=(240, 200))
+
+Pad img with white pixels to height:width ratio `sz[1]:sz[2]`.
+
+Note that `sz` here is not the output size.
+"""
+function center_pad(img::AbstractMatrix, sz=(240, 200))
+ # the default size (240, 200) is used in DemoCards
+ fillvalue = oneunit(eltype(img))
+
+ # make sure we don't shrink the image
+ h, w = size(img)
+ ratio = sz[1]/sz[2]
+ pad_sz = h/w > ratio ? (h, round(Int, w / ratio)) : (round(Int, h * ratio), w)
+ pad_sz = max.(size(img), pad_sz)
+
+ offset = (pad_sz .- size(img)) .÷ 2
+ PaddedView(fillvalue, img, ntuple(i -> -offset[i]:pad_sz[i]-offset[i]+1, ndims(img)))
+end
+
+function drawborder(img, fillvalue=colorant"pink")
+ img = copy(img)
+ img[1, 1:end] .= fillvalue
+ img[1:end, 1] .= fillvalue
+ img[end, 1:end] .= fillvalue
+ img[1:end, end] .= fillvalue
+ img
+end
diff --git a/docs/operations/config.json b/docs/operations/config.json
new file mode 100644
index 00000000..3acad879
--- /dev/null
+++ b/docs/operations/config.json
@@ -0,0 +1,9 @@
+{
+ "order": [
+ "affine",
+ "distortions",
+ "size",
+ "misc"
+ ]
+}
+
\ No newline at end of file
diff --git a/docs/operations/distortions/config.json b/docs/operations/distortions/config.json
new file mode 100644
index 00000000..36de087a
--- /dev/null
+++ b/docs/operations/distortions/config.json
@@ -0,0 +1,7 @@
+{
+ "title": "Distortions",
+ "order": [
+ "elasticdistortion.jl"
+ ],
+ "description": "Aside from affine transformations, Augmentor also provides functionality for performing a variety of distortions. These types of operations usually provide a much larger distribution of possible output images."
+}
diff --git a/docs/operations/distortions/elasticdistortion.jl b/docs/operations/distortions/elasticdistortion.jl
new file mode 100644
index 00000000..e0e6db29
--- /dev/null
+++ b/docs/operations/distortions/elasticdistortion.jl
@@ -0,0 +1,32 @@
+# ---
+# title: ElasticDistortion
+# cover: elasticdistortion.gif
+# id: op_elastic
+# ---
+
+# Smoothed random distortion
+
+using Augmentor
+using ImageShow, ImageCore
+using Random
+
+img_in = testpattern(RGB, ratio=0.5)
+
+mosaicview(
+ img_in,
+ augment(img_in, ElasticDistortion(15,15,0.1)),
+ augment(img_in, ElasticDistortion(10,10,0.2,4,3,true));
+ fillvalue=colorant"white", nrow=1, npad=10
+)
+
+# ## Reference
+
+#md # ```@docs
+#md # ElasticDistortion
+#md # ```
+
+## save covers #src
+using ImageMagick #src
+include(joinpath("..", "assets", "utilities.jl")) #src
+cover = make_gif(testpattern(RGB, ratio=0.5), ElasticDistortion(15,15,0.1), 10) #src
+ImageMagick.save("elasticdistortion.gif", cover; fps=2) #src
diff --git a/docs/operations/index.md b/docs/operations/index.md
new file mode 100644
index 00000000..16e3b8ed
--- /dev/null
+++ b/docs/operations/index.md
@@ -0,0 +1,11 @@
+# [Supported Operations](@id operations)
+
+Augmentor provides a wide variety of build-in image operations.
+This page provides an overview of all exported operations
+organized by their main category. These categories are chosen
+because they serve some practical purpose. For example Affine
+Operations allow for a special optimization under the hood when
+chained together.
+
+
+{{{democards}}}
diff --git a/docs/operations/misc/config.json b/docs/operations/misc/config.json
new file mode 100644
index 00000000..74737941
--- /dev/null
+++ b/docs/operations/misc/config.json
@@ -0,0 +1,6 @@
+{
+ "order":[
+ "layout.jl",
+ "utilities.jl"
+ ]
+}
diff --git a/docs/operations/misc/layout.jl b/docs/operations/misc/layout.jl
new file mode 100644
index 00000000..fc042651
--- /dev/null
+++ b/docs/operations/misc/layout.jl
@@ -0,0 +1,51 @@
+# ---
+# title: Colorant conversion and channel layout
+# cover: layout.gif
+# description: a set of commonly used basic operations that wrapped by Augmentor
+# ---
+
+# Augmentor has warpped some commonly used basic operations so that you can use to build the augmentation
+# pipeline. The `internal` column is what you'd probably do outside of `Augmentor`.
+
+# | Category | internal | Augmentor |
+# | --- | --- | --- |
+# | Conversion | `T.(img)` | `ConvertEltype(T)` |
+# | Information Layout | `ImageCore.channelview` | `SplitChannels` |
+# | Information Layout | `ImageCore.colorview` | `CombineChannels` |
+# | Information Layout | `Base.permutedims` | `PermuteDims` |
+# | Information Layout | `Base.reshape` | `Reshape` |
+
+# It is not uncommon that machine learning frameworks require the data in a specific form and layout.
+# For example many deep learning frameworks expect the colorchannel of the images to be encoded in
+# the third dimension of a 4-dimensional array. Augmentor allows to convert from (and to) these
+# different layouts using special operations that are mainly useful in the beginning or end of a
+# augmentation pipeline.
+
+using Augmentor
+using ImageCore
+
+## 300×400 Matrix{RGB{N0f8}, 2} => 300×400×3 Array{Float32, 3}
+img = testpattern(RGB, ratio=0.5)
+img_in = augment(img, SplitChannels() |> PermuteDims(2, 3, 1) |> ConvertEltype(Float32))
+
+## 300×400×3 Array{Float32, 3} => 300×400 Matrix{RGB{N0f8}, 2}
+img_out = augment(img_in, ConvertEltype(N0f8) |> PermuteDims(3, 1, 2) |> CombineChannels(RGB))
+
+img_out == img
+
+
+# ## References
+
+#md # ```@docs
+#md # ConvertEltype
+#md # SplitChannels
+#md # CombineChannels
+#md # PermuteDims
+#md # Reshape
+#md # ```
+
+## save covers #src
+using ImageMagick #src
+include(joinpath("..", "assets", "utilities.jl")) #src
+cover = make_gif(testpattern(RGB, ratio=0.5), ConvertEltype(Gray{N0f8}), 2) #src
+ImageMagick.save("layout.gif", cover; fps=1) #src
diff --git a/docs/operations/misc/utilities.jl b/docs/operations/misc/utilities.jl
new file mode 100644
index 00000000..e38d2a78
--- /dev/null
+++ b/docs/operations/misc/utilities.jl
@@ -0,0 +1,33 @@
+# ---
+# title: Composition utilities
+# cover: utilities.gif
+# description: a set of helper opeartions that may be useful when compositing more complex augmentation workflow
+# ---
+
+# Aside from "true" operations that specify some kind of transformation, there are also a couple of
+# special utility operations used for functionality such as stochastic branching.
+
+using Augmentor
+using Random
+Random.seed!(1337)
+
+img_in = testpattern(RGB, ratio=0.5)
+img_out = augment(img_in, Either(0.5=>NoOp(), 0.25=>FlipX(), 0.25=>FlipY()))
+#md nothing #hide
+
+# 
+
+# ## References
+
+#md # ```@docs
+#md # NoOp
+#md # Either
+#md # CacheImage
+#md # ```
+
+## save covers #src
+using ImageMagick #src
+include(joinpath("..", "assets", "utilities.jl")) #src
+pl = Either(ShearX(-5:5), ShearY(-5:5)) |> Rotate(-10:10) |> Either(NoOp(), FlipX(), FlipY()) |> CropNative(axes(img_in)) #src
+cover = make_gif(img_in, pl, 10) #src
+ImageMagick.save("utilities.gif", cover; fps=2) #src
diff --git a/docs/operations/size/config.json b/docs/operations/size/config.json
new file mode 100644
index 00000000..5cd5b418
--- /dev/null
+++ b/docs/operations/size/config.json
@@ -0,0 +1,9 @@
+{
+ "title": "Resizing and Subsetting",
+ "order": [
+ "crop.jl",
+ "cropsize.jl",
+ "cropratio.jl"
+ ],
+ "description": "The process of cropping is useful to discard parts of the input image. To provide this functionality lazily, applying a crop introduces a layer of representation called a \"view\" or SubArray. This is different yet compatible with how affine operations or other special purpose implementations work. This means that chaining a crop with some affine operation is perfectly fine if done sequentially. However, it is generally not advised to combine affine operations with crop operations within an [`Either`](@ref) block. Doing that would force the [`Either`](@ref) to trigger the eager computation of its branches in order to preserve type-stability."
+}
diff --git a/docs/operations/size/crop.jl b/docs/operations/size/crop.jl
new file mode 100644
index 00000000..d6c0b08c
--- /dev/null
+++ b/docs/operations/size/crop.jl
@@ -0,0 +1,58 @@
+# ---
+# title: Crop
+# cover: crop.gif
+# ---
+
+# Subset image using `Crop` and `CropNative`
+
+using Augmentor
+using ImageShow, ImageCore
+using OffsetArrays
+
+img_in = testpattern(RGB, ratio=0.5)
+img_out = augment(img_in, Crop(20:75,25:120))
+
+mosaicview(img_in, img_out; fillvalue=colorant"white", nrow=1)
+
+# If the input image is plain arrays without offset indices, then `Crop` and `CropNative` is equivalent.
+
+augment(img_in, Crop(20:75,25:120)) == augment(img_in, CropNative(20:75,25:120))
+
+# Whether you should use `Crop` or `CropNative` depends on if you want to take the index offset
+# of the input image into consideration.
+
+imgo_in = OffsetArray(img_in, -50, -50)
+imgo_out = augment(imgo_in, Crop(20:75,25:120))
+imgo_out_native = augment(imgo_in, CropNative(20:75,25:120))
+
+(
+ imgo_in[(first.(axes(imgo_in)) .+ (20, 25))...] == imgo_out[1, 1],
+ imgo_in[20, 25] == imgo_out_native[1, 1]
+)
+
+
+# A typical scenario that you may want to use `CropNative` is when you have affine operations, e.g.,
+# `Rotate` and `ShearX`.
+
+mosaicview(
+ augment(img_in, Rotate(30) |> Crop(axes(img_in))),
+ augment(img_in, Rotate(30) |> CropNative(axes(img_in))),
+
+ augment(img_in, ShearX(10) |> Crop(axes(img_in))),
+ augment(img_in, ShearX(10) |> CropNative(axes(img_in)));
+
+ fillvalue=colorant"white", rowmajor=true, nrow=2, npad=10
+)
+
+# ## Reference
+
+#md # ```@docs
+#md # Crop
+#md # CropNative
+#md # ```
+
+## save covers #src
+using ImageMagick #src
+include(joinpath("..", "assets", "utilities.jl")) #src
+cover = make_gif(img_in, Crop(20:75,25:120), 2) #src
+ImageMagick.save("crop.gif", cover; fps=1) #src
diff --git a/docs/operations/size/cropratio.jl b/docs/operations/size/cropratio.jl
new file mode 100644
index 00000000..08279685
--- /dev/null
+++ b/docs/operations/size/cropratio.jl
@@ -0,0 +1,37 @@
+# ---
+# title: CropRatio
+# cover: cropratio.gif
+# ---
+
+# Crop centered window to fit given aspect ratio
+
+using Augmentor
+using ImageShow, ImageCore
+
+img_in = testpattern(RGB, ratio=0.5)
+img_out = augment(img_in, CropRatio()) # crop out a square window
+
+mosaicview(img_in, img_out; nrow=1)
+
+# `RCropRatio` is a random version that randomly choose a crop center -- not necessarily the center
+# of the input image.
+
+augment(img_in, RCropRatio())
+#md nothing #hide
+
+# 
+
+# ## Reference
+
+#md # ```@docs
+#md # CropRatio
+#md # RCropRatio
+#md # ```
+
+
+## save covers #src
+using ImageMagick #src
+using FileIO #src
+include(joinpath("..", "assets", "utilities.jl")) #src
+cover = make_gif(img_in, RCropRatio(), 5) #src
+ImageMagick.save("cropratio.gif", cover; fps=1) #src
diff --git a/docs/operations/size/cropsize.jl b/docs/operations/size/cropsize.jl
new file mode 100644
index 00000000..582bcbee
--- /dev/null
+++ b/docs/operations/size/cropsize.jl
@@ -0,0 +1,37 @@
+# ---
+# title: CropSize
+# cover: cropsize.gif
+# ---
+
+# Crop centered window to given size
+
+using Augmentor
+using ImageShow, ImageCore
+
+img_in = testpattern(RGB, ratio=0.5)
+img_out = augment(img_in, CropSize(70, 70)) # crop out a square window
+
+mosaicview(img_in, img_out; nrow=1, npad=10)
+
+# `RCropSize` is a random version that randomly choose a crop center -- not necessarily the center
+# of the input image.
+
+augment(img_in, CropSize(70, 70))
+#md nothing #hide
+
+# 
+
+# ## Reference
+
+#md # ```@docs
+#md # CropSize
+#md # RCropSize
+#md # ```
+
+
+## save covers #src
+using ImageMagick #src
+using FileIO #src
+include(joinpath("..", "assets", "utilities.jl")) #src
+cover = make_gif(testpattern(RGB, ratio=0.5), RCropSize(70, 70), 5) #src
+ImageMagick.save("cropsize.gif", cover; fps=1) #src
diff --git a/docs/optable.jl b/docs/optable.jl
deleted file mode 100644
index 4f3bc9dc..00000000
--- a/docs/optable.jl
+++ /dev/null
@@ -1,101 +0,0 @@
-# This file is imported by most Augmentor.Operations
-# documentation pages. The purpose of this file is to generate
-# one or more example images for what the operation does.
-# Additionally the result is returned as a markdown table
-# showing the input image and the corresponding output image.
-#
-# Example use for single output image as png:
-#
-# ```@eval
-# include("optable.jl")
-# @optable Rotate(15)
-# ```
-#
-# Example use for 8 output images compiled into a gif:
-#
-# ```@eval
-# include("optable.jl")
-# @optable 8 => Rotate(-15:15)
-# ```
-
-using Augmentor, Images, Colors
-using Reel, PaddedViews, OffsetArrays
-Reel.set_output_type("gif")
-
-srand(1337)
-
-if !isfile("../assets/testpattern.png")
- pattern = imresize(testpattern(), (240, 320))
- save("../assets/testpattern.png", pattern)
-end
-
-pattern = load("../assets/testpattern.png")
-pattern_noalpha = ((1 .- alpha.(pattern)) .* colorant"#F3F6F6") .+ (alpha.(pattern) .* color.(pattern))
-
-function drawborder!(img, col)
- img[1:end, 1] .= fill(col, size(img,1))
- img[1:end, end] .= fill(col, size(img,1))
- img[1, 1:end] .= fill(col, size(img,2))
- img[end, 1:end] .= fill(col, size(img,2))
- img
-end
-
-centered(img) = OffsetArray(img, convert(Tuple, 1 .- round.(Int, ImageTransformations.center(img))))
-
-macro optable(expr)
- if expr.args[1] == :(=>) && expr.args[2] isa Int
- n = expr.args[2]
- nexpr = expr.args[3]
- name = string(nexpr.args[1])
- descr = string(nexpr)
- :(optable($(esc(nexpr)), $name, $descr, $n))
- elseif expr.args[1] == :(=>) && expr.args[2] isa String
- name = expr.args[2]
- nexpr = expr.args[3]
- descr = string(nexpr)
- :(optable($(esc(nexpr)), $name, $descr))
- else
- name = string(expr.args[1])
- descr = string(expr)
- :(optable($(esc(expr)), $name, $descr))
- end
-end
-
-function optable(op, name, descr)
- fname = joinpath("..", "assets", string(name, ".png"))
- i = 2
- while isfile(fname)
- fname = joinpath("..", "assets", string(name, i, ".png"))
- i = i + 1
- end
- out = augment(pattern, op)
- save(fname, out)
- header = length(descr) < 20 ? "Output for `$descr`" : "`$descr`"
- tbl = string(
- "Input | $header\n",
- "------|--------\n",
- " | \n"
- )
- Markdown.parse(tbl)
-end
-
-function optable(op, name, descr, n)
- fname = joinpath("..", "assets", string(name, ".gif"))
- i = 2
- while isfile(fname)
- fname = joinpath("..", "assets", string(name, i, ".gif"))
- i = i + 1
- end
- raw_imgs = [centered(drawborder!(augment(pattern_noalpha, op), colorant"pink")) for i in 1:n]
- imgs = map(parent, map(copy, [paddedviews(colorant"#F3F6F6", raw_imgs...)...]))
- insert!(imgs, 1, first(imgs)) # otherwise loop isn't smooth
- film = roll(imgs, fps = 2)
- write(fname, film)
- header = length(descr) < 20 ? "Samples for `$descr`" : "`$descr`"
- tbl = string(
- "Input | $header\n",
- "------|--------\n",
- " | \n"
- )
- Markdown.parse(tbl)
-end
diff --git a/docs/src/LICENSE.md b/docs/src/LICENSE.md
index 9b6b98e3..cc6d1c22 100644
--- a/docs/src/LICENSE.md
+++ b/docs/src/LICENSE.md
@@ -1,5 +1,6 @@
# LICENSE
```@eval
-Markdown.parse_file(joinpath(@__DIR__, "../LICENSE.md"))
+using Markdown, Augmentor
+Markdown.parse_file(joinpath(pkgdir(Augmentor), "LICENSE.md"))
```
diff --git a/docs/src/gettingstarted.md b/docs/src/gettingstarted.md
index 17c8095f..0930b6fd 100644
--- a/docs/src/gettingstarted.md
+++ b/docs/src/gettingstarted.md
@@ -21,7 +21,7 @@ in the case you would like to contribute to the package, you can
manually choose to be on the latest (untagged) version.
```julia
-Pkg.checkout("Augmentor")
+Pkg.develop("Augmentor")
```
## Example
@@ -35,7 +35,7 @@ can be downloaded
[here](https://isic-archive.com/api/v1/image/5592ac599fc3c13155a57a85/thumbnail)
using their [Web API](https://isic-archive.com/api/v1).
-```julia-repl
+```julia
julia> using Augmentor, ISICArchive
julia> img = get(ImageThumbnailRequest(id = "5592ac599fc3c13155a57a85"))
@@ -62,7 +62,9 @@ julia> img_new = augment(img, pl)
```
```@eval
-using Augmentor, ISICArchive;
+using Augmentor, ISICArchive
+using ImageCore, ImageMagick
+using Random
img = get(ImageThumbnailRequest(id = "5592ac599fc3c13155a57a85"))
@@ -73,34 +75,29 @@ pl = Either(1=>FlipX(), 1=>FlipY(), 2=>NoOp()) |>
Zoom(1:0.05:1.2) |>
Resize(64, 64)
-img_new = augment(img, pl)
-
-using Plots
-pyplot(reuse = true)
-default(bg_outside=colorant"#F3F6F6")
-srand(123)
-
-# Create image that shows the input
-plot(img, size=(256,169), xlim=(1,255), ylim=(1,168), grid=false, ticks=true)
-Plots.png(joinpath("assets","isic_in.png"))
-
-# create animate gif that shows 10 outputs
-anim = @animate for i=1:10
- plot(augment(img, pl), size=(169,169), xlim=(1,63), ylim=(1,63), grid=false, ticks=true)
+# modified from operations/assets/gif.jl
+function make_gif(img, pl, num_sample; post_op=identity, random_seed=1337)
+ Random.seed!(random_seed)
+
+ fillvalue = oneunit(eltype(img))
+ frames = sym_paddedviews(
+ fillvalue,
+ post_op(img),
+ [post_op(augment(img, pl)) for _ in 1:num_sample-1]...
+ )
+ cat(frames..., dims=3)
end
-Plots.gif(anim, joinpath("assets","isic_out.gif"), fps = 2)
+
+ImageMagick.save(joinpath("assets","isic_in.png"), img)
+preview = make_gif(img, pl, 10)[:, :, 2:end]
+ImageMagick.save(joinpath("assets", "isic_out.gif"), preview; fps=2)
nothing
```
The function `augment` will generate a single augmented image
from the given input image and pipeline. To visualize the effect
-we compiled a few resulting output images into a GIF using the
-plotting library
-[Plots.jl](https://github.com/JuliaPlots/Plots.jl) with the
-[PyPlot.jl](https://github.com/JuliaPy/PyPlot.jl) back-end.
-You can inspect the full code by clicking on "Edit on Github" in
-the top right corner of this page.
+we compiled a few resulting output images into a GIF.
Input (`img`) | | Output (`img_new`)
:---------------------------:|:-:|:------------------------------:
diff --git a/docs/src/images.md b/docs/src/images.md
index 9c02ec6f..8f5ea10c 100644
--- a/docs/src/images.md
+++ b/docs/src/images.md
@@ -18,7 +18,7 @@ new community members. This beginner's guide is an attempt to
provide a step-by-step overview of how pixel data is handled in
Julia. To get a more detailed explanation on some particular
concept involved, please take a look at the documentation of the
-[JuliaImages](https://juliaimages.github.io) ecosystem.
+[JuliaImages](https://juliaimages.org/) ecosystem.
## Multi-dimensional Arrays
@@ -49,7 +49,7 @@ representation.
```jldoctest 1
julia> memory = [0x1, 0x2, 0x3, 0x4, 0x5, 0x6]
-6-element Array{UInt8,1}:
+6-element Vector{UInt8}:
0x01
0x02
0x03
@@ -61,11 +61,11 @@ julia> memory = [0x1, 0x2, 0x3, 0x4, 0x5, 0x6]
The same block of memory could also be interpreted differently.
For example we could think of this as a matrix with 3 rows and 2
columns instead (or even the other way around). The function
-`reinterpret` allows us to do just that
+`reshape` allows us to do just that
```jldoctest 1
-julia> A = reinterpret(UInt8, memory, (3,2))
-3×2 Array{UInt8,2}:
+julia> A = reshape(memory, (3, 2))
+3×2 Matrix{UInt8}:
0x01 0x04
0x02 0x05
0x03 0x06
@@ -95,7 +95,7 @@ This idea can also be generalized for higher dimensions. For
example we can think of this as a 3D array as well.
```jldoctest 1
-julia> reinterpret(UInt8, memory, (3,1,2))
+julia> reshape(memory, (3, 1, 2))
3×1×2 Array{UInt8,3}:
[:, :, 1] =
0x01
@@ -115,7 +115,7 @@ any number of practically empty dimensions, otherwise known as
*singleton dimensions*.
```jldoctest 1
-julia> reinterpret(UInt8, memory, (3,1,1,1,2))
+julia> reshape(memory, (3,1,1,1,2))
3×1×1×1×2 Array{UInt8,5}:
[:, :, 1, 1, 1] =
0x01
@@ -153,8 +153,8 @@ If for some reason that is not the case there are two possible
ways to convert the image to that format.
```jldoctest 1
-julia> At = reinterpret(UInt8, memory, (3,2))' # "row-major" layout
-2×3 Array{UInt8,2}:
+julia> At = collect(reshape(memory, (3,2))') # "row-major" layout
+2×3 Matrix{UInt8}:
0x01 0x02 0x03
0x04 0x05 0x06
```
@@ -166,22 +166,19 @@ julia> At = reinterpret(UInt8, memory, (3,2))' # "row-major" layout
```jldoctest 1
julia> B = permutedims(At, (2,1))
- 3×2 Array{UInt8,2}:
+ 3×2 Matrix{UInt8}:
0x01 0x04
0x02 0x05
0x03 0x06
```
-2. The second way is using the function
- `ImageCore.permuteddimsview` which results in a lazy view that
+2. The second way is using `Base.PermutedDimsArray` which results in a lazy view that
does not allocate a new array but instead only computes the
correct values when queried.
```jldoctest 1
- julia> using ImageCore
-
- julia> C = permuteddimsview(At, (2,1))
- 3×2 PermutedDimsArray(::Array{UInt8,2}, (2, 1)) with element type UInt8:
+ julia> C = PermutedDimsArray(At, (2,1))
+ 3×2 PermutedDimsArray(::Matrix{UInt8}, (2, 1)) with eltype UInt8:
0x01 0x04
0x02 0x05
0x03 0x06
@@ -204,7 +201,7 @@ consider our original vector `memory` again.
```jldoctest 1
julia> memory = [0x1, 0x2, 0x3, 0x4, 0x5, 0x6]
-6-element Array{UInt8,1}:
+6-element Vector{UInt8}:
0x01
0x02
0x03
@@ -219,7 +216,7 @@ vector of 3 `UInt16` elements.
```jldoctest 1
julia> reinterpret(UInt16, memory)
-3-element Array{UInt16,1}:
+3-element reinterpret(UInt16, ::Vector{UInt8}):
0x0201
0x0403
0x0605
@@ -274,7 +271,7 @@ above, and interpret the underlying memory as a vector of to
```julia-repl
julia> reinterpret(MyRGB, memory)
-2-element Array{MyRGB,1}:
+2-element Vector{MyRGB}:
MyRGB(0x01,0x02,0x03)
MyRGB(0x04,0x05,0x06)
```
@@ -321,7 +318,7 @@ numbers still depends on the number of underlying bits in the
memory, but that is not much of an issue.
```jldoctest 1
-julia> using FixedPointNumbers;
+julia> using ImageCore; # ImageCore reexports FixedPointNumbers and Colors
julia> reinterpret(N0f8, 0xFF)
1.0N0f8
@@ -355,15 +352,13 @@ per color channel, and with the second command as a single pixel
of 16 bit per color channel
```jldoctest 1
-julia> using Colors, FixedPointNumbers;
-
julia> reinterpret(RGB{N0f8}, memory)
-2-element Array{RGB{N0f8},1}:
+2-element reinterpret(RGB{N0f8}, ::Vector{UInt8}):
RGB{N0f8}(0.004,0.008,0.012)
RGB{N0f8}(0.016,0.02,0.024)
julia> reinterpret(RGB{N0f16}, memory)
-1-element Array{RGB{N0f16},1}:
+1-element reinterpret(RGB{N0f16}, ::Vector{UInt8}):
RGB{N0f16}(0.00783,0.01567,0.02351)
```
diff --git a/docs/src/index.md b/docs/src/index.md
index 8b5b9a5c..eb7dd356 100644
--- a/docs/src/index.md
+++ b/docs/src/index.md
@@ -29,10 +29,22 @@ first few examples of the [MNIST
database](http://yann.lecun.com/exdb/mnist/).
```@eval
-# I can't use Reel.jl, because the way it stores the tmp pngs
-# causes the images to be upscaled too much.
-using Augmentor, MLDatasets, Images, Colors
-using PaddedViews, OffsetArrays
+using Augmentor, ImageCore, ImageMagick
+using MLDatasets
+using Random
+
+# copied from operations/assets/gif.jl
+function make_gif(img, pl, num_sample; random_seed=1337, kwargs...)
+ fillvalue = oneunit(eltype(img[1]))
+
+ init_frame = mosaicview(img; kwargs...)
+ frames = map(1:num_sample-1) do _
+ mosaicview(map(x->augment(x, pl), img)...; kwargs...)
+ end
+
+ frames = sym_paddedviews(fillvalue, init_frame, frames...)
+ cat(frames..., dims=3)
+end
pl = ElasticDistortion(6, scale=0.3, border=true) |>
Rotate([10, -5, -3, 0, 3, 5, 10]) |>
@@ -40,28 +52,16 @@ pl = ElasticDistortion(6, scale=0.3, border=true) |>
CropSize(28, 28) |>
Zoom(0.9:0.1:1.2)
-md_imgs = String[]
-for i in 1:24
- srand(i) # somehow srand in the beginning isn't enough
- input = MNIST.convert2image(MNIST.traintensor(i))
- imgs = [augment(input, pl) for j in 1:20]
- insert!(imgs, 1, first(imgs)) # otherwise loop isn't smooth
- fnames = map(imgs) do img
- tpath = tempname() * ".png"
- save(tpath, img)
- tpath
- end
- args = reduce(vcat, [[fname, "-delay", "1x4", "-alpha", "deactivate"] for fname in fnames])
- convert = strip(readstring(`which convert`))
- outname = joinpath("assets", "idx_mnist_$i.gif")
- run(`$convert $args $outname`)
- push!(md_imgs, "[](@ref mnist)")
- foreach(fname -> rm(fname), fnames)
-end
-Markdown.parse(join(md_imgs, " "))
+n_samples, n_frames = 24, 10
+imgs = [MNIST.convert2image(MNIST.traintensor(i)) for i in 1:n_samples]
+preview = make_gif(imgs, pl, n_frames; nrow=1)
+
+ImageMagick.save("mnist_preview.gif", RGB(1, 1, 1) .- preview; fps=3)
```
-The Julia version of Augmentor is engineered specifically for
+
+
+The Julia version of **Augmentor** is engineered specifically for
high performance applications. It makes use of multiple
heuristics to generate efficient tailor-made code for the
concrete user-specified augmentation pipeline. In particular
@@ -69,6 +69,8 @@ Augmentor tries to avoid the need for any intermediate images,
but instead aims to compute the output image directly from the
input in one single pass.
+For the Python version of Augmentor, you can find it [here](https://github.com/mdbloice/Augmentor)
+
## Where to begin?
If this is the first time you consider using Augmentor.jl for
@@ -81,10 +83,6 @@ Pages = ["gettingstarted.md"]
Depth = 2
```
-**Augmentor.jl** is the [Julia](https://julialang.org) package
-for Augmentor. You can find the Python version
-[here](https://github.com/mdbloice/Augmentor).
-
## Introduction and Motivation
If you are new to image augmentation in general, or are simply
@@ -102,7 +100,7 @@ free to browse the following documents for a crash course on how
image data is represented in the Julia language, as well as how
to visualize it. For more information on image processing in
Julia, take a look at the documentation for the vast
-[`JuliaImages`](https://juliaimages.github.io/latest/) ecosystem.
+[`JuliaImages`](https://juliaimages.github.io/stable/) ecosystem.
```@contents
Pages = ["images.md"]
diff --git a/docs/src/interface.md b/docs/src/interface.md
index 5443f719..2056a7c7 100644
--- a/docs/src/interface.md
+++ b/docs/src/interface.md
@@ -30,9 +30,9 @@ detail.
```
Depending on the complexity of your problem, you may want to
-iterate between `2.` and `3.` to identify an appropriate
+iterate between step `2.` and `3.` to identify an appropriate
pipeline.
-Take a look at the [Elastic Distortions Tutorial](@ref elastic)
+Take a look at the [Elastic Distortions Tutorial](@ref mnist_elastic)
for an example of how such an iterative process could look like.
## [Defining a Pipeline](@id pipeline)
@@ -50,7 +50,7 @@ them down to the biggest possible square, and lastly resize the
image(s) to a fixed size of 64 by 64 pixel. Such a pipeline would
be defined as follows:
-```julia-repl
+```jldoctest; setup = :(using Augmentor)
julia> pl = Rotate(14) |> CropRatio(1) |> Resize(64,64)
3-step Augmentor.ImmutablePipeline:
1.) Rotate 14 degree
diff --git a/docs/src/operations.md b/docs/src/operations.md
deleted file mode 100644
index 08eabd5c..00000000
--- a/docs/src/operations.md
+++ /dev/null
@@ -1,129 +0,0 @@
-```@eval
-using Augmentor, Images, Colors
-srand(1337)
-pattern = imresize(restrict(restrict(testpattern())), (60, 80))
-save("assets/tiny_pattern.png", pattern)
-# Affine Transformations
-save("assets/tiny_FlipX.png", augment(pattern, FlipX()))
-save("assets/tiny_FlipY.png", augment(pattern, FlipY()))
-save("assets/tiny_Rotate90.png", augment(pattern, Rotate90()))
-save("assets/tiny_Rotate270.png", augment(pattern, Rotate270()))
-save("assets/tiny_Rotate180.png", augment(pattern, Rotate180()))
-save("assets/tiny_Rotate.png", augment(pattern, Rotate(15)))
-save("assets/tiny_ShearX.png", augment(pattern, ShearX(10)))
-save("assets/tiny_ShearY.png", augment(pattern, ShearY(10)))
-save("assets/tiny_Scale.png", augment(pattern, Scale(0.9,1.2)))
-save("assets/tiny_Zoom.png", augment(pattern, Zoom(0.9,1.2)))
-# Distortions
-srand(1337)
-save("assets/tiny_ED1.png", augment(pattern, ElasticDistortion(15,15,0.1)))
-save("assets/tiny_ED2.png", augment(pattern, ElasticDistortion(10,10,0.2,4,3,true)))
-# Resizing and Subsetting
-save("assets/tiny_Resize.png", augment(pattern, Resize(60,60)))
-save("assets/tiny_Crop.png", augment(pattern, Rotate(45) |> Crop(1:50,1:80)))
-save("assets/tiny_CropNative.png", augment(pattern, Rotate(45) |> CropNative(1:50,1:80)))
-save("assets/tiny_CropSize.png", augment(pattern, CropSize(20,65)))
-save("assets/tiny_CropRatio.png", augment(pattern, CropRatio(1)))
-srand(1337)
-save("assets/tiny_RCropRatio.png", augment(pattern, RCropRatio(1)))
-# Conversion
-save("assets/tiny_ConvertEltype.png", augment(pattern, ConvertEltype(GrayA{N0f8})))
-nothing;
-```
-
-# [Supported Operations](@id operations)
-
-Augmentor provides a wide varitey of build-in image operations.
-This page provides an overview of all exported operations
-organized by their main category. These categories are chosen
-because they serve some practical purpose. For example Affine
-Operations allow for a special optimization under the hood when
-chained together.
-
-!!! tip
-
- Click on an image operation for more details.
-
-## Affine Transformations
-
-A sizeable amount of the provided operations fall under the
-category of **affine transformations**. As such, they can be
-described using what is known as an [affine
-map](https://en.wikipedia.org/wiki/Affine_transformation), which
-are inherently compose-able if chained together. However,
-utilizing such a affine formulation requires (costly)
-interpolation, which may not always be needed to achieve the
-desired effect. For that reason do some of the operations below
-also provide a special purpose implementation to produce their
-specified result. Those are usually preferred over the affine
-formulation if sensible considering the complete pipeline.
-
-| **Input** | | **[`FlipX`](@ref FlipX)** | **[`FlipY`](@ref FlipY)** | **[`Rotate90`](@ref Rotate90)** | **[`Rotate270`](@ref Rotate270)** | **[`Rotate180`](@ref Rotate180)** |
-|:---------:|:--:|:-------------------:|:-------------------:|:----------------------:|:-----------------------:|:-----------------------:|
-|  | → | [](@ref FlipX) | [](@ref FlipY) | [](@ref Rotate90) | [](@ref Rotate270) | [](@ref Rotate180) |
-| **Input** | | **[`Rotate`](@ref Rotate)** | **[`ShearX`](@ref ShearX)** | **[`ShearY`](@ref ShearY)** | **[`Scale`](@ref Scale)** | **[`Zoom`](@ref Zoom)** |
-|  | → | [](@ref Rotate) | [](@ref ShearX) | [](@ref ShearY) | [](@ref Scale) | [](@ref Zoom) |
-
-## Distortions
-
-Aside from affine transformations, Augmentor also provides
-functionality for performing a variety of distortions. These
-types of operations usually provide a much larger distribution of
-possible output images.
-
-| **Input** | | **[`ElasticDistortion`](@ref ElasticDistortion)** |
-|:---------:|:--:|:-------------------------------------------------:|
-|  | → | [](@ref ElasticDistortion) |
-
-## Resizing and Subsetting
-
-The input images from a given dataset can be of various shapes
-and sizes. Yet, it is often required by the algorithm that the
-data must be of uniform structure. To that end Augmentor provides
-a number of ways to alter or subset given images.
-
-| **Input** | | **[`Resize`](@ref Resize)** |
-|:---------:|:--:|:---------------------------:|
-|  | → | [](@ref Resize) |
-
-The process of cropping is useful to discard parts of the input
-image. To provide this functionality lazily, applying a crop
-introduces a layer of representation called a "view" or
-`SubArray`. This is different yet compatible with how affine
-operations or other special purpose implementations work. This
-means that chaining a crop with some affine operation is
-perfectly fine if done sequentially. However, it is generally not
-advised to combine affine operations with crop operations within
-an [`Either`](@ref) block. Doing that would force the
-[`Either`](@ref) to trigger the eager computation of its branches
-in order to preserve type-stability.
-
-| **Input** | | **[`Crop`](@ref Crop)** | **[`CropNative`](@ref CropNative)** | **[`CropSize`](@ref CropSize)** | **[`CropRatio`](@ref CropRatio)** | **[`RCropRatio`](@ref RCropRatio)** |
-|:---------:|:--:|:------------------:|:------------------------:|:----------------------:|:-----------------------:|:------------------------:|
-|  | → | [](@ref Crop) | [](@ref CropNative) | [](@ref CropSize) | [](@ref CropRatio) | [](@ref RCropRatio) |
-
-## Element-wise Transformations and Layout
-
-It is not uncommon that machine learning frameworks require the
-data in a specific form and layout. For example many deep
-learning frameworks expect the colorchannel of the images to be
-encoded in the third dimension of a 4-dimensional array.
-Augmentor allows to convert from (and to) these different layouts
-using special operations that are mainly useful in the beginning
-or end of a augmentation pipeline.
-
-Category | Available Operations
-----------------------|-----------------------------------------------
-Conversion | [`ConvertEltype`](@ref ConvertEltype) (e.g. convert to grayscale)
-Mapping | [`MapFun`](@ref MapFun), [`AggregateThenMapFun`](@ref AggregateThenMapFun)
-Information Layout | [`SplitChannels`](@ref SplitChannels), [`CombineChannels`](@ref CombineChannels), [`PermuteDims`](@ref PermuteDims), [`Reshape`](@ref Reshape)
-
-## Utility Operations
-
-Aside from "true" operations that specify some kind of
-transformation, there are also a couple of special utility
-operations used for functionality such as stochastic branching.
-
-Category | Available Operations
-----------------------|-----------------------------------------------
-Utility Operations | [`NoOp`](@ref NoOp), [`CacheImage`](@ref CacheImage), [`Either`](@ref Either)
diff --git a/docs/src/operations/aggmapfun.md b/docs/src/operations/aggmapfun.md
deleted file mode 100644
index b6608764..00000000
--- a/docs/src/operations/aggmapfun.md
+++ /dev/null
@@ -1,5 +0,0 @@
-# [AggregateThenMapFun: Aggregate and Map over Image](@id AggregateThenMapFun)
-
-```@docs
-AggregateThenMapFun
-```
diff --git a/docs/src/operations/cacheimage.md b/docs/src/operations/cacheimage.md
deleted file mode 100644
index 6a76f55c..00000000
--- a/docs/src/operations/cacheimage.md
+++ /dev/null
@@ -1,5 +0,0 @@
-# [CacheImage: Buffer current state](@id CacheImage)
-
-```@docs
-CacheImage
-```
diff --git a/docs/src/operations/combinechannels.md b/docs/src/operations/combinechannels.md
deleted file mode 100644
index cb971660..00000000
--- a/docs/src/operations/combinechannels.md
+++ /dev/null
@@ -1,5 +0,0 @@
-# [ComineChannels: Combine color channels](@id CombineChannels)
-
-```@docs
-CombineChannels
-```
diff --git a/docs/src/operations/converteltype.md b/docs/src/operations/converteltype.md
deleted file mode 100644
index ae94e80e..00000000
--- a/docs/src/operations/converteltype.md
+++ /dev/null
@@ -1,10 +0,0 @@
-# [ConvertEltype: Color conversion](@id ConvertEltype)
-
-```@docs
-ConvertEltype
-```
-
-```@eval
-include("optable.jl")
-@optable ConvertEltype(GrayA{N0f8})
-```
diff --git a/docs/src/operations/crop.md b/docs/src/operations/crop.md
deleted file mode 100644
index 2ef9c8dc..00000000
--- a/docs/src/operations/crop.md
+++ /dev/null
@@ -1,10 +0,0 @@
-# [Crop: Subset image](@id Crop)
-
-```@docs
-Crop
-```
-
-```@eval
-include("optable.jl")
-@optable Crop(70:140,25:155)
-```
diff --git a/docs/src/operations/cropnative.md b/docs/src/operations/cropnative.md
deleted file mode 100644
index b2447c26..00000000
--- a/docs/src/operations/cropnative.md
+++ /dev/null
@@ -1,17 +0,0 @@
-# [CropNative: Subset image](@id CropNative)
-
-```@docs
-CropNative
-```
-
-```@eval
-include("optable.jl")
-@optable "cropn1" => (Rotate(45),Crop(1:210,1:280))
-@optable "cropn2" => (Rotate(45),CropNative(1:210,1:280))
-tbl = string(
- "`(Rotate(45), Crop(1:210,1:280))` | `(Rotate(45), CropNative(1:210,1:280))`\n",
- "-----|-----\n",
- " | \n"
-)
-Markdown.parse(tbl)
-```
diff --git a/docs/src/operations/cropratio.md b/docs/src/operations/cropratio.md
deleted file mode 100644
index b32fb61e..00000000
--- a/docs/src/operations/cropratio.md
+++ /dev/null
@@ -1,10 +0,0 @@
-# [CropRatio: Crop centered window](@id CropRatio)
-
-```@docs
-CropRatio
-```
-
-```@eval
-include("optable.jl")
-@optable CropRatio(1)
-```
diff --git a/docs/src/operations/cropsize.md b/docs/src/operations/cropsize.md
deleted file mode 100644
index bd1393d1..00000000
--- a/docs/src/operations/cropsize.md
+++ /dev/null
@@ -1,10 +0,0 @@
-# [CropSize: Crop centered window](@id CropSize)
-
-```@docs
-CropSize
-```
-
-```@eval
-include("optable.jl")
-@optable CropSize(45,225)
-```
diff --git a/docs/src/operations/either.md b/docs/src/operations/either.md
deleted file mode 100644
index d4697a1c..00000000
--- a/docs/src/operations/either.md
+++ /dev/null
@@ -1,5 +0,0 @@
-# [Either: Stochastic branches](@id Either)
-
-```@docs
-Either
-```
diff --git a/docs/src/operations/elasticdistortion.md b/docs/src/operations/elasticdistortion.md
deleted file mode 100644
index e74101ef..00000000
--- a/docs/src/operations/elasticdistortion.md
+++ /dev/null
@@ -1,15 +0,0 @@
-# [ElasticDistortion: Smoothed random distortions](@id ElasticDistortion)
-
-```@docs
-ElasticDistortion
-```
-
-```@eval
-include("optable.jl")
-@optable 10 => ElasticDistortion(15,15,0.1)
-```
-
-```@eval
-include("optable.jl")
-@optable 10 => ElasticDistortion(10,10,0.2,4,3,true)
-```
diff --git a/docs/src/operations/flipx.md b/docs/src/operations/flipx.md
deleted file mode 100644
index d527c2e9..00000000
--- a/docs/src/operations/flipx.md
+++ /dev/null
@@ -1,10 +0,0 @@
-# [FlipX: Mirror horizontally](@id FlipX)
-
-```@docs
-FlipX
-```
-
-```@eval
-include("optable.jl")
-@optable FlipX()
-```
diff --git a/docs/src/operations/flipy.md b/docs/src/operations/flipy.md
deleted file mode 100644
index 4f9ddf7d..00000000
--- a/docs/src/operations/flipy.md
+++ /dev/null
@@ -1,10 +0,0 @@
-# [FlipY: Mirror vertically](@id FlipY)
-
-```@docs
-FlipY
-```
-
-```@eval
-include("optable.jl")
-@optable FlipY()
-```
diff --git a/docs/src/operations/mapfun.md b/docs/src/operations/mapfun.md
deleted file mode 100644
index 4a66e415..00000000
--- a/docs/src/operations/mapfun.md
+++ /dev/null
@@ -1,5 +0,0 @@
-# [MapFun: Map function over Image](@id MapFun)
-
-```@docs
-MapFun
-```
diff --git a/docs/src/operations/noop.md b/docs/src/operations/noop.md
deleted file mode 100644
index 404c8890..00000000
--- a/docs/src/operations/noop.md
+++ /dev/null
@@ -1,5 +0,0 @@
-# [NoOp: Identity function](@id NoOp)
-
-```@docs
-NoOp
-```
diff --git a/docs/src/operations/permutedims.md b/docs/src/operations/permutedims.md
deleted file mode 100644
index 02c691b4..00000000
--- a/docs/src/operations/permutedims.md
+++ /dev/null
@@ -1,5 +0,0 @@
-# [PermuteDims: Change dimension order](@id PermuteDims)
-
-```@docs
-PermuteDims
-```
diff --git a/docs/src/operations/rcropratio.md b/docs/src/operations/rcropratio.md
deleted file mode 100644
index 544ba287..00000000
--- a/docs/src/operations/rcropratio.md
+++ /dev/null
@@ -1,10 +0,0 @@
-# [RCropRatio: Crop random window](@id RCropRatio)
-
-```@docs
-RCropRatio
-```
-
-```@eval
-include("optable.jl")
-@optable 10 => RCropRatio(1)
-```
diff --git a/docs/src/operations/reshape.md b/docs/src/operations/reshape.md
deleted file mode 100644
index 28e1b71b..00000000
--- a/docs/src/operations/reshape.md
+++ /dev/null
@@ -1,5 +0,0 @@
-# [Reshape: Reinterpret shape](@id Reshape)
-
-```@docs
-Reshape
-```
diff --git a/docs/src/operations/resize.md b/docs/src/operations/resize.md
deleted file mode 100644
index 57a51c05..00000000
--- a/docs/src/operations/resize.md
+++ /dev/null
@@ -1,10 +0,0 @@
-# [Resize: Set static image size](@id Resize)
-
-```@docs
-Resize
-```
-
-```@eval
-include("optable.jl")
-@optable Resize(100,150)
-```
diff --git a/docs/src/operations/rotate.md b/docs/src/operations/rotate.md
deleted file mode 100644
index c21791eb..00000000
--- a/docs/src/operations/rotate.md
+++ /dev/null
@@ -1,25 +0,0 @@
-# [Rotate: Arbitrary rotations](@id Rotate)
-
-```@docs
-Rotate
-```
-
-In contrast to the special case rotations outlined above, the
-type `Rotate` can describe any arbitrary number of degrees. It
-will always perform the rotation around the center of the image.
-This can be particularly useful when combining the operation with
-[`CropNative`](@ref).
-
-```@eval
-include("optable.jl")
-@optable Rotate(15)
-```
-
-It is also possible to pass some abstract vector to the
-constructor, in which case Augmentor will randomly sample one of
-its elements every time the operation is applied.
-
-```@eval
-include("optable.jl")
-@optable 10 => Rotate(-10:10)
-```
diff --git a/docs/src/operations/rotate180.md b/docs/src/operations/rotate180.md
deleted file mode 100644
index 48743f47..00000000
--- a/docs/src/operations/rotate180.md
+++ /dev/null
@@ -1,10 +0,0 @@
-# [Rotate180: Rotate by 180 degree](@id Rotate180)
-
-```@docs
-Rotate180
-```
-
-```@eval
-include("optable.jl")
-@optable Rotate180()
-```
diff --git a/docs/src/operations/rotate270.md b/docs/src/operations/rotate270.md
deleted file mode 100644
index 71eaba1b..00000000
--- a/docs/src/operations/rotate270.md
+++ /dev/null
@@ -1,10 +0,0 @@
-# [Rotate270: Rotate downwards 90 degree](@id Rotate270)
-
-```@docs
-Rotate270
-```
-
-```@eval
-include("optable.jl")
-@optable Rotate270()
-```
diff --git a/docs/src/operations/rotate90.md b/docs/src/operations/rotate90.md
deleted file mode 100644
index f13f42c8..00000000
--- a/docs/src/operations/rotate90.md
+++ /dev/null
@@ -1,10 +0,0 @@
-# [Rotate90: Rotate upwards 90 degree](@id Rotate90)
-
-```@docs
-Rotate90
-```
-
-```@eval
-include("optable.jl")
-@optable Rotate90()
-```
diff --git a/docs/src/operations/scale.md b/docs/src/operations/scale.md
deleted file mode 100644
index 216fba96..00000000
--- a/docs/src/operations/scale.md
+++ /dev/null
@@ -1,27 +0,0 @@
-# [Scale: Relative resizing](@id Scale)
-
-```@docs
-Scale
-```
-```@eval
-include("optable.jl")
-@optable Scale(0.9,0.5)
-```
-
-In the case that only a single scale factor is specified, the
-operation will assume that the intention is to scale all
-dimensions uniformly by that factor.
-
-```@eval
-include("optable.jl")
-@optable Scale(1.2)
-```
-
-It is also possible to pass some abstract vector(s) to the
-constructor, in which case Augmentor will randomly sample one of
-its elements every time the operation is applied.
-
-```@eval
-include("optable.jl")
-@optable 10 => Scale(0.9:0.05:1.2)
-```
diff --git a/docs/src/operations/shearx.md b/docs/src/operations/shearx.md
deleted file mode 100644
index 1ef08993..00000000
--- a/docs/src/operations/shearx.md
+++ /dev/null
@@ -1,23 +0,0 @@
-# [ShearX: Shear horizontally](@id ShearX)
-
-```@docs
-ShearX
-```
-
-It will always perform the transformation around the center of
-the image. This can be particularly useful when combining the
-operation with [`CropNative`](@ref).
-
-```@eval
-include("optable.jl")
-@optable ShearX(10)
-```
-
-It is also possible to pass some abstract vector to the
-constructor, in which case Augmentor will randomly sample one of
-its elements every time the operation is applied.
-
-```@eval
-include("optable.jl")
-@optable 10 => ShearX(-10:10)
-```
diff --git a/docs/src/operations/sheary.md b/docs/src/operations/sheary.md
deleted file mode 100644
index 6732e25a..00000000
--- a/docs/src/operations/sheary.md
+++ /dev/null
@@ -1,23 +0,0 @@
-# [ShearY: Shear vertically](@id ShearY)
-
-```@docs
-ShearY
-```
-
-It will always perform the transformation around the center of
-the image. This can be particularly useful when combining the
-operation with [`CropNative`](@ref).
-
-```@eval
-include("optable.jl")
-@optable ShearY(10)
-```
-
-It is also possible to pass some abstract vector to the
-constructor, in which case Augmentor will randomly sample one of
-its elements every time the operation is applied.
-
-```@eval
-include("optable.jl")
-@optable 10 => ShearY(-10:10)
-```
diff --git a/docs/src/operations/splitchannels.md b/docs/src/operations/splitchannels.md
deleted file mode 100644
index ad25e434..00000000
--- a/docs/src/operations/splitchannels.md
+++ /dev/null
@@ -1,5 +0,0 @@
-# [SplitChannels: Separate color channels](@id SplitChannels)
-
-```@docs
-SplitChannels
-```
diff --git a/docs/src/operations/zoom.md b/docs/src/operations/zoom.md
deleted file mode 100644
index 00fcf22b..00000000
--- a/docs/src/operations/zoom.md
+++ /dev/null
@@ -1,18 +0,0 @@
-# [Zoom: Scale without resize](@id Zoom)
-
-```@docs
-Zoom
-```
-```@eval
-include("optable.jl")
-@optable Zoom(1.2)
-```
-
-It is also possible to pass some abstract vector to the
-constructor, in which case Augmentor will randomly sample one of
-its elements every time the operation is applied.
-
-```@eval
-include("optable.jl")
-@optable 10 => Zoom(0.9:0.05:1.3)
-```
diff --git a/examples/mnist_elastic.jl b/examples/mnist_elastic.jl
deleted file mode 100644
index 90247b73..00000000
--- a/examples/mnist_elastic.jl
+++ /dev/null
@@ -1,142 +0,0 @@
-#' # [MNIST: Elastic Distortions](@id elastic)
-
-#' In this example we are going to use Augmentor on the famous
-#' **MNIST database of handwritten digits** [^MNIST1998] to
-#' reproduce the elastic distortions discussed in [^SIMARD2003].
-#' It may be interesting to point out, that the way Augmentor
-#' implements distortions is a little different to how it is
-#' described by the authors of the paper.
-#' This is for a couple of reasons, most notably that we want the
-#' parameters for our deformations to be independent of the size
-#' of image it is applied on. As a consequence the
-#' parameter-numbers specified in the paper are not 1-to-1
-#' transferable to Augmentor.
-
-#' If the effects are sensible for the dataset, then applying
-#' elastic distortions can be a really effective way to improve
-#' the generalization ability of the network.
-#' That said, our implementation of [`ElasticDistortion`](@ref)
-#' has a lot of possible parameters to choose from. To that end,
-#' we will introduce a simple strategy for interactively
-#' exploring the parameter space on our dataset of interest.
-
-#md #' !!! note
-#md #'
-#md #' This tutorial was designed to be performed in a
-#md #' [Juypter](https://jupyter.org/) notebook. You can
-#md #' find a link to the Juypter version of this tutorial
-#md #' in the top right corner of this page.
-
-#' ## Loading the MNIST Trainingset
-
-#' In order to access and visualize the MNIST images we employ
-#' the help of two additional Julia packages. In the interest of
-#' time and space we will not go into great detail about their
-#' functionality. Feel free to click on their respective names to
-#' find out more information about the utility they can provide.
-#'
-#' - [Images.jl](https://github.com/JuliaImages/Images.jl) will
-#' provide us with the necessary tools for working with image
-#' data in Julia.
-#'
-#' - [MLDatasets.jl](https://github.com/JuliaML/MLDatasets.jl)
-#' has an MNIST submodule that offers a convenience interface
-#' to read the MNIST database.
-
-#' The function `MNIST.traintensor` returns the MNIST training
-#' images corresponding to the given indices as a
-#' multi-dimensional array. These images are stored in the native
-#' horizontal-major memory layout as a single floating point
-#' array, where all values are scaled to be between 0.0 and 1.0.
-
-using Images, MLDatasets
-train_tensor = MNIST.traintensor()
-@show summary(train_tensor);
-#md nothing # hide
-
-#' This horizontal-major format is the standard way of utilizing
-#' this dataset for training machine learning models.
-#' In this tutorial, however, we are more interested in working
-#' with the MNIST images as actual Julia images in vertical-major
-#' layout, and as black digits on white background.
-
-#' We can convert the "tensor" to a `Colorant` array using the
-#' provided function `MNIST.convert2image`.
-#' This way, Julia knows we are dealing with image data and can
-#' tell programming environments such as Juypter how to visualize
-#' it. If you are working in the terminal you may want to use the
-#' package [ImageInTerminal.jl](https://github.com/JuliaImages/ImageInTerminal.jl)
-
-train_images = MNIST.convert2image(train_tensor)
-img_1 = train_images[:,:,1] # show first image
-#md save("mnist_1.png",repeat(img_1,inner=(4,4))) # hide
-#md nothing # hide
-
-#md #' 
-
-#' ## Visualizing the Effects
-
-#' Before applying an operation (or pipeline of operations) on
-#' some dataset to train a network, we strongly recommend
-#' investing some time in selecting a decent set of hyper
-#' parameters for the operation(s). A useful tool for tasks like
-#' this is the package [Interact.jl](https://github.com/JuliaGizmos/Interact.jl).
-#' We will use this package to define a number of widgets for
-#' controlling the parameters to our operation.
-
-#' Note that while the code below only focuses on configuring
-#' the parameters of a single operation, specifically
-#' [`ElasticDistortion`](@ref), it could also be adapted to tweak
-#' a whole pipeline. Take a look at the corresponding section in
-#' [High-level Interface](@ref pipeline) for more information
-#' on how to define and use a pipeline.
-
-# These two package will provide us with the capabilities
-# to perform interactive visualisations in a jupyter notebook
-using Augmentor, Interact, Reactive
-
-# The manipulate macro will turn the parameters of the
-# loop into interactive widgets.
-@manipulate for
- unpaused = true,
- ticks = fpswhen(signal(unpaused), 5.),
- image_index = 1:100,
- grid_size = 3:20,
- scale = .1:.1:.5,
- sigma = 1:5,
- iterations = 1:6,
- free_border = true
- op = ElasticDistortion(grid_size, grid_size, # equal width & height
- sigma = sigma,
- scale = scale,
- iter = iterations,
- border = free_border)
- augment(train_images[:, :, image_index], op)
-end
-#md nothing # hide
-
-#md #' Executing the code above in a Juypter notebook will result
-#md #' in the following interactive visualisation. You can now
-#md #' use the sliders to investigate the effects that different
-#md #' parameters have on the MNIST training images.
-#md #'
-#md #' !!! tip
-#md #'
-#md #' You should always use your **training** set to do this
-#md #' kind of visualisation (not the test test!). Otherwise
-#md #' you are likely to achieve overly optimistic (i.e. biased)
-#md #' results during training.
-#md #'
-#md #' 
-
-#' Congratulations! With just a few simple lines of code, you
-#' created a simple interactive tool to visualize your image
-#' augmentation pipeline. Once you found a set of parameters that
-#' you think are appropriate for your dataset you can go ahead
-#' and train your model.
-
-#' ## References
-#'
-#' [^MNIST1998]: LeCun, Yan, Corinna Cortes, Christopher J.C. Burges. ["The MNIST database of handwritten digits"](http://yann.lecun.com/exdb/mnist/) Website. 1998.
-#'
-#' [^SIMARD2003]: Simard, Patrice Y., David Steinkraus, and John C. Platt. ["Best practices for convolutional neural networks applied to visual document analysis."](https://www.microsoft.com/en-us/research/publication/best-practices-for-convolutional-neural-networks-applied-to-visual-document-analysis/) ICDAR. Vol. 3. 2003.
diff --git a/examples/mnist_knet.jl b/examples/mnist_knet.jl
deleted file mode 100644
index 8c8b6d9e..00000000
--- a/examples/mnist_knet.jl
+++ /dev/null
@@ -1,577 +0,0 @@
-#' # MNIST: Knet.jl CNN
-info("MNIST: Knet.jl CNN example") #jl-only
-
-#' In this tutorial we will adapt the
-#' [MNIST example](http://denizyuret.github.io/Knet.jl/latest/tutorial.html#Convolutional-neural-network-1)
-#' from [Knet.jl](https://github.com/denizyuret/Knet.jl)
-#' to utilize a custom augmentation pipeline.
-#' In order to showcase the effect that image augmentation can
-#' have on a neural network's ability to generalize, we will
-#' limit the training set to just the first 500 images (of the
-#' available 60,000!). For more information on the dataset see
-#' [^MNIST1998].
-
-#md #' !!! note
-#md #'
-#md #' This tutorial is also available as a
-#md #' [Juypter](https://jupyter.org/) notebook. You can
-#md #' find a link to the Juypter version of this tutorial
-#md #' in the top right corner of this page.
-
-#' ## Preparing the MNIST dataset
-info("Preparing the MNIST dataset") #jl-only
-
-#' In order to access, prepare, and visualize the MNIST images we
-#' employ the help of three additional Julia packages. In the
-#' interest of time and space we will not go into great detail
-#' about their functionality. Feel free to click on their
-#' respective names to find out more information about the
-#' utility they can provide.
-#'
-#' - [MLDatasets.jl](https://github.com/JuliaML/MLDatasets.jl)
-#' has an MNIST submodule that offers a convenience interface
-#' to read the MNIST database.
-#'
-#' - [Images.jl](https://github.com/JuliaImages/Images.jl) will
-#' provide us with the necessary tools to process and display
-#' the image data in Julia / Juypter.
-#'
-#' - [MLDataUtils.jl](https://github.com/JuliaML/MLDataUtils.jl)
-#' implements a variety of functions to convert and partition
-#' Machine Learning datasets. This will help us prepare the
-#' MNIST data to be used with Knet.jl.
-
-using Images, MLDatasets, MLDataUtils
-srand(42);
-#md nothing # hide
-
-#' As you may have seen previously in the
-#' [elastic distortions tutorial](@ref elastic), the function
-#' `MNIST.traintensor` returns the MNIST training images
-#' corresponding to the given indices as a multi-dimensional
-#' array. These images are stored in the native horizontal-major
-#' memory layout as a single array. Because we specify that
-#' the `eltype` of that array should be `Float32`, all the
-#' individual values are scaled to be between `0.0` and `1.0`.
-#' Also note, how the observations are laid out along the last
-#' array dimension
-
-@show summary(MNIST.traintensor(Float32, 1:500));
-#md nothing # hide
-
-#' The corresponding label of each image is stored as an integer
-#' value between `0` and `9`. That means that if the label has
-#' the value `3`, then the corresponding image is known to be a
-#' handwritten "3". To show a more concrete example, the
-#' following code reveals that the first training image denotes a
-#' "5" and the second training image a "0" (etc).
-
-@show summary(MNIST.trainlabels(1:500))
-println("First eight labels: ", join(MNIST.trainlabels(1:8),", "))
-
-#' For Knet we will require a slightly format for the images
-#' and also the labels. More specifically, we add an additional
-#' singleton dimension of length 1 to our image array. Think of
-#' this as our single color channel (because MNIST images are gray).
-#' Additionally we will convert our labels to proper 1-based indices.
-#' This is because some functions provided by Knet expect the labels
-#' to be in this format. We will do all this by creating a little
-#' utility function that we will name `prepare_mnist`.
-
-"""
- prepare_mnist(images, labels) -> (X, Y)
-
-Change the dimension layout x1×x2×N of the given array
-`images` to x1×x2×1×N and return the result as `X`.
-The given integer vector `labels` is transformed into
-an integer vector denoting 1-based class indices.
-"""
-function prepare_mnist(images, labels)
- X = reshape(images, (28, 28, 1, :))
- Y = convertlabel(LabelEnc.Indices{Int8}, labels, 0:9)
- X, Y
-end
-#md nothing # hide
-
-#' With `prepare_mnist` defined, we can now use it in conjunction
-#' with the functions in the `MLDatasets.MNIST` sub-module to load
-#' and prepare our training set. Recall that for this tutorial only
-#' the first 500 images of the training set will be used.
-
-train_x, train_y = prepare_mnist(MNIST.traintensor(Float32, 1:500), MNIST.trainlabels(1:500))
-@show summary(train_x) summary(train_y);
-[MNIST.convert2image(train_x[:,:,1,i]) for i in 1:8]
-#md tmp = hcat(ans...) # hide
-#md save("mnist_knet_train.png",repeat(tmp, inner=(4,4))) # hide
-#md nothing # hide
-
-#md #' 
-
-#' Similarly, we use `MNIST.testtensor` and `MNIST.testlabels`
-#' to load the full MNIST test set. We will utilize that data to
-#' measure how well the network is able to generalize with and
-#' without augmentation.
-
-test_x, test_y = prepare_mnist(MNIST.testtensor(Float32), MNIST.testlabels())
-@show summary(test_x) summary(test_y);
-[MNIST.convert2image(test_x[:,:,1,i]) for i in 1:8]
-#md tmp = hcat(ans...) # hide
-#md save("mnist_knet_test.png",repeat(tmp, inner=(4,4))) # hide
-#md nothing # hide
-
-#md #' 
-
-#' ## Defining the Network
-info("Defining the Network") #jl-only
-
-#' With the dataset prepared, we can now define and instantiate
-#' our neural network. To keep things simple, we will use the same
-#' convolutional network as defined in the
-#' [MNIST example](http://denizyuret.github.io/Knet.jl/latest/tutorial.html#Convolutional-neural-network-1)
-#' of the Knet.jl package.
-
-using Knet
-#md nothing # hide
-
-#' The first thing we will do is define the forward pass through
-#' the network. This will effectively outline the computation
-#' graph of the network architecture. Note how this does not
-#' define some details, such as the number of neurons per layer.
-#' We will define those later when initializing our
-#' vector of weight arrays `w`.
-
-"""
- forward(w, x) -> a
-
-Compute the forward pass for the given minibatch `x` by using the
-neural network parameters in `w`. The resulting (unnormalized)
-activations of the last layer are returned as `a`.
-"""
-function forward(w, x)
- # conv1 (2x2 maxpool)
- a1 = pool(relu.(conv4(w[1], x) .+ w[2]))
- # conv2 (2x2 maxpool)
- a2 = pool(relu.(conv4(w[3], a1) .+ w[4]))
- # dense1 (relu)
- a3 = relu.(w[5] * mat(a2) .+ w[6])
- # dense2 (identity)
- a4 = w[7] * a3 .+ w[8]
- return a4
-end
-#md nothing # hide
-
-#' In order to be able to train our network we need to choose a
-#' cost function. Because this is a classification problem we will
-#' use the negative log-likelihood (provided by `Knet.nll`).
-#' With the cost function defined we can the simply use the
-#' higher-order function `grad` to create a new function `costgrad`
-#' that computes us the corresponding gradients.
-
-"""
- cost(w, x, y) -> AbstractFloat
-
-Compute the per-instance negative log-likelihood for the data
-in the minibatch `(x, y)` given the network with the current
-parameters in `w`.
-"""
-cost(w, x, y) = nll(forward(w, x), y)
-costgrad = grad(cost)
-#md nothing # hide
-
-#' Aside from the cost function that we need for training, we
-#' would also like a more interpretable performance measurement.
-#' In this tutorial we will use "accuracy" for its simplicity
-#' and because we know that the class distribution for MNIST
-#' is close to uniform.
-
-"""
- acc(w, X, Y; [batchsize]) -> Float64
-
-Compute the accuracy for the data in `(X,Y)` given the network
-with the current parameters in `w`. The resulting value is
-computed by iterating over the data in minibatches of size
-`batchsize`.
-"""
-function acc(w, X, Y; batchsize = 100)
- sum = 0; count = 0
- for (x_cpu, y) in eachbatch((X, Y), maxsize = batchsize)
- x = KnetArray{Float32}(x_cpu)
- sum += Int(accuracy(forward(w,x), y, average = false))
- count += length(y)
- end
- return sum / count
-end
-#md nothing # hide
-
-#' Before we can train or even just use our network, we need to
-#' define how we initialize `w`, which is our the vector of
-#' parameter arrays. The dimensions of these individual arrays
-#' specify the filter sizes and number of neurons.
-#' It can be helpful to compare the indices here with the indices
-#' used in our `forward` function to see which array corresponds
-#' to which computation node of our network.
-
-function weights(atype = KnetArray{Float32})
- w = Array{Any}(8)
- # conv1
- w[1] = xavier(5,5,1,20)
- w[2] = zeros(1,1,20,1)
- # conv2
- w[3] = xavier(5,5,20,50)
- w[4] = zeros(1,1,50,1)
- # dense1
- w[5] = xavier(500,800)
- w[6] = zeros(500,1)
- # dense2
- w[7] = xavier(10,500)
- w[8] = zeros(10,1)
- return map(a->convert(atype,a), w)
-end
-#md nothing # hide
-
-
-#' ## Training without Augmentation
-info("Training baseline network without augmentation") #jl-only
-
-#' In order to get an intuition for how useful augmentation can
-#' be, we need a sensible baseline to compare to. To that end, we
-#' will first train the network we just defined using only the
-#' (unaltered) 500 training examples.
-
-#' The package
-#' [ValueHistories.jl](https://github.com/JuliaML/ValueHistories.jl)
-#' will help us record the accuracy during the training process.
-#' We will use those logs later to visualize the differences
-#' between having augmentation or no augmentation.
-
-using ValueHistories
-using ProgressMeter #jl-only
-
-#' To keep things simple, we will not overly optimize our
-#' training function. Thus, we will be content with using a
-#' closure. Because both, the baseline and the augmented version,
-#' will share this "inefficiency", we should still get a decent
-#' enough picture of their performance differences.
-
-function train_baseline(; epochs = 500, batchsize = 100, lr = .03)
- w = weights()
- log = MVHistory()
- p = Progress(epochs, desc = "Baseline: ") #jl-only
- for epoch in 1:epochs
- for (batch_x_cpu, batch_y) in eachbatch((train_x ,train_y), batchsize)
- batch_x = KnetArray{Float32}(batch_x_cpu)
- g = costgrad(w, batch_x, batch_y)
- Knet.update!(w, g, lr = lr)
- end
-
- next!(p) #jl-only
- if (epoch % 5) == 0
- train = acc(w, train_x, train_y)
- test = acc(w, test_x, test_y)
- @trace log epoch train test
- msg = "epoch " * lpad(epoch,4) * ": train accuracy " * rpad(round(train,3),5,"0") * ", test accuracy " * rpad(round(test,3),5,"0")
- cancel(p, msg, :blue) #jl-only
-#md println(msg)
-#jp println(msg)
- end
- end
- finish!(p) #jl-only
- log
-end
-#md nothing # hide
-
-#' Aside from the accuracy, we will also keep an eye on the
-#' training time. In particular we would like to see if and how
-#' the addition of augmentation causes our training time to
-#' increase.
-
-train_baseline(epochs=1) # warm-up
-baseline_log = @time train_baseline(epochs=200);
-#md nothing # hide
-
-#' As we can see, the accuracy on the training set is around a
-#' 100%, while the accuracy on the test set peaks around 90%. For
-#' a mere 500 training examples, this isn't actually that bad of
-#' a result.
-
-#' ## Integrating Augmentor
-info("Training network with augmentation") #jl-only
-
-#' Now that we have a network architecture with a baseline to
-#' compare to, let us finally see what it takes to add Augmentor
-#' to our experiment. First, we need to include the package to
-#' our experiment.
-
-using Augmentor
-
-#' The next step, and maybe the most human-hour consuming part of
-#' adding image augmentation to a prediction problem, is to
-#' design and select a sensible augmentation pipeline. Take a
-#' look at the [elastic distortions tutorial](@ref elastic) for
-#' an example of how to do just that.
-
-#' For this example, we already choose a quite complicated but
-#' promising augmentation pipeline for you. This pipeline was
-#' designed to yield a large variation of effects as well as to
-#' showcase how even deep pipelines are quite efficient in terms
-#' of performance.
-
-pl = Reshape(28,28) |>
- PermuteDims(2,1) |>
- ShearX(-5:5) * ShearY(-5:5) |>
- Rotate(-15:15) |>
- CropSize(28,28) |>
- Zoom(0.9:0.1:1.2) |>
- CacheImage() |>
- ElasticDistortion(10) |>
- PermuteDims(2,1) |>
- Reshape(28,28,1)
-println(pl) #jl-only
-
-#' Most of the used operations are quite self explanatory, but
-#' there are some details about this pipeline worth pointing out
-#' explicitly.
-
-#' 1. We use the operation [`PermuteDims`](@ref) to convert the
-#' horizontal-major MNIST image to a julia-native
-#' vertical-major image. The vertical-major image is then
-#' processed and converted back to a horizontal-major array.
-#' We mainly do this here to showcase the option, but it is
-#' also to keep consistent with how the data is usually used
-#' in the literature. Alternatively, one could just work with
-#' the MNIST data in a vertical-major format all the way
-#' through without any issue.
-
-#' 2. As counter-intuitive as it sounds, the operation
-#' [`CacheImage`](@ref) right before
-#' [`ElasticDistortion`](@ref) is actually used to improve
-#' performance. If we were to omit it, then the whole pipeline
-#' would be applied in one single pass. In this case, applying
-#' distortions on top of affine transformations lazily is in
-#' fact less efficient than using a temporary variable.
-
-#' With the pipeline now defined, let us quickly peek at what
-#' kind of effects we can achieve with it. In particular, lets
-#' apply the pipeline multiple times to the first training image
-#' and look at what kind of results it produces.
-
-#jp [MNIST.convert2image(reshape(augment(train_x[:,:,:,1], pl), (28, 28))) for i in 1:2, j in 1:8]
-#md [MNIST.convert2image(reshape(augment(train_x[:,:,:,1], pl), (28, 28))) for i in 1:8, j in 1:2]
-#md tmp = vcat(hcat(ans[:,1]...), hcat(ans[:,2]...)) # hide
-#md save("mnist_knet_aug.png",repeat(tmp, inner=(4,4))) # hide
-#md nothing # hide
-
-#md #' 
-
-#' As we can see, we can achieve a wide range of effects, from
-#' more subtle to more pronounced. The important part is that all
-#' examples are still clearly representative of the true label.
-
-#' Next, we have to adapt the function `train_baseline` to make
-#' use of our augmentation pipeline. To integrate Augmentor
-#' efficiently, there are three necessary changes we have to
-#' make.
-
-#' 1. Preallocate a buffer with the same size and element type
-#' that each batch has.
-#'
-#' ```
-#' batch_x_aug = zeros(Float32, 28, 28, 1, batchsize)
-#' ```
-
-#' 2. Add a call to [`augmentbatch!`](@ref) in the inner loop of
-#' the batch iterator using our pipeline and buffer.
-#'
-#' ```
-#' augmentbatch!(batch_x_aug, batch_x_org, pl)
-#' ```
-
-#' 3. Replace `batch_x_org` with `batch_x_aug` in the constructor
-#' of `KnetArray`.
-#'
-#' ```
-#' batch_x = KnetArray{Float32}(batch_x_aug)
-#' ```
-
-
-#' Applying these changes to our `train_baseline` function
-#' will give us something similar to the following function.
-#' Note how all the other parts of the function remain exactly
-#' the same as before.
-
-function train_augmented(; epochs = 500, batchsize = 100, lr = .03)
- w = weights()
- log = MVHistory()
- p = Progress(epochs, desc = "Augmented: ") #jl-only
- batch_x_aug = zeros(Float32, size(train_x,1), size(train_x,2), 1, batchsize)
- for epoch in 1:epochs
- for (batch_x_cpu, batch_y) in eachbatch((train_x ,train_y), batchsize)
- augmentbatch!(CPUThreads(), batch_x_aug, batch_x_cpu, pl)
- batch_x = KnetArray{Float32}(batch_x_aug)
- g = costgrad(w, batch_x, batch_y)
- Knet.update!(w, g, lr = lr)
- end
-
- next!(p) #jl-only
- if (epoch % 5) == 0
- train = acc(w, train_x, train_y)
- test = acc(w, test_x, test_y)
- @trace log epoch train test
- msg = "epoch " * lpad(epoch,4) * ": train accuracy " * rpad(round(train,3),5,"0") * ", test accuracy " * rpad(round(test,3),5,"0")
- cancel(p, msg, :blue) #jl-only
-#md println(msg)
-#jp println(msg)
- end
- end
- finish!(p) #jl-only
- log
-end
-#md nothing # hide
-
-#' You may have noticed in the code above that we also pass a
-#' `CPUThreads()` as the first argument to [`augmentbatch!`](@ref).
-#' This instructs Augmentor to process the images of the batch in
-#' parallel using multi-threading. For this to work properly you
-#' will need to set the environment variable `JULIA_NUM_THREADS`
-#' to the number of threads you wish to use. You can check how
-#' many threads are used with the function `Threads.nthreads()`
-
-@show Threads.nthreads();
-#md nothing # hide
-
-#' Now that all pieces are in place, let us train our network
-#' once more. We will use the same parameters except that now
-#' instead of the original training images we will be using
-#' randomly augmented images. This will cause every epoch to be
-#' different.
-
-train_augmented(epochs=1) # warm-up
-augmented_log = @time train_augmented(epochs=200);
-#md nothing # hide
-
-#' As we can see, our network reaches far better results on our
-#' testset than our baseline network did. However, we can also
-#' see that the training took quite a bit longer than before.
-#' This difference generally decreases as the complexity of the
-#' utilized neural network increases. Yet another way to improve
-#' performance (aside from simplifying the augmentation pipeline)
-#' would be to increase the number of available threads.
-
-#' ## Improving Performance
-info("Improving Performance") #jl-only
-
-#' One of the most effective ways to make the most out of the
-#' available resources is to augment the next (couple) mini-batch
-#' while the current minibatch is being processed on the GPU.
-#' We can do this via julia's build in parallel computing
-#' capabilities
-
-#' First we need a worker process that will be responsible for
-#' augmenting our dataset each epoch. This worker also needs
-#' access to a couple of our packages
-
-# addprocs(1)
-# @everywhere using Augmentor, MLDataUtils
-
-#' Next, we replace the inner `eachbatch` loop with a more
-#' complicated version using a `RemoteChannel` to exchange and
-#' queue the augmented data.
-
-function async_train_augmented(; epochs = 500, batchsize = 100, lr = .03)
- w = weights()
- log = MVHistory()
- p = Progress(epochs, desc = "Async Augmented: ") #jl-only
- for epoch in 1:epochs
- @sync begin
- local_ch = Channel{Tuple}(4) # prepare up to 4 minibatches in adavnce
- remote_ch = RemoteChannel(()->local_ch)
- @spawn begin
- # This block is executed on the worker process
- batch_x_aug = zeros(Float32, size(train_x,1), size(train_x,2), 1, batchsize)
- for (batch_x_cpu, batch_y) in eachbatch((train_x ,train_y), batchsize)
- # we are still using multithreading
- augmentbatch!(CPUThreads(), batch_x_aug, batch_x_cpu, pl)
- put!(remote_ch, (batch_x_aug, batch_y))
- end
- close(remote_ch)
- end
- @async begin
- # This block is executed on the main process
- for (batch_x_aug, batch_y) in local_ch
- batch_x = KnetArray{Float32}(batch_x_aug)
- g = costgrad(w, batch_x, batch_y)
- Knet.update!(w, g, lr = lr)
- end
- end
- end
-
- next!(p) #jl-only
- if (epoch % 5) == 0
- train = acc(w, train_x, train_y)
- test = acc(w, test_x, test_y)
- @trace log epoch train test
- msg = "epoch " * lpad(epoch,4) * ": train accuracy " * rpad(round(train,3),5,"0") * ", test accuracy " * rpad(round(test,3),5,"0")
- cancel(p, msg, :blue) #jl-only
-#md println(msg)
-#jp println(msg)
- end
- end
- finish!(p) #jl-only
- log
-end
-#md nothing # hide
-
-#' Note that for this toy example the overhead of this approach
-#' is greater than the benefit.
-
-#' ## Visualizing the Results
-info("Visualizing the Results") #jl-only
-
-#' Before we end this tutorial, let us make use the
-#' [Plots.jl](https://github.com/JuliaPlots/Plots.jl) package to
-#' visualize and discuss the recorded training curves.
-#' We will plot the accuracy curves of both networks side by side
-#' in order to get a good feeling about their differences.
-
-using Plots
-#jp pyplot()
-#md pyplot()
-#md nothing # hide
-unicodeplots() #jl-only
-
-#+
-
-#md default(bg_outside=colorant"#FFFFFF") # hide
-plt = plot(
- plot(baseline_log, title="Baseline", ylim=(.5,1)),
- plot(augmented_log, title="Augmented", ylim=(.5,1)),
- size = (900, 400),
- xlab = "Epoch",
- ylab = "Accuracy",
- markersize = 1
-)
-#jp plt
-#md png(plt, "mnist_knet_curves.png") # hide
-#md nothing # hide
-
-#md #' 
-
-#' Note how the accuracy on the (unaltered) training set
-#' increases faster for the baseline network than for the
-#' augmented one. This is to be expected, since our augmented
-#' network doesn't actually use the unaltered images for
-#' training, and thus has not actually seen them. Given this
-#' information, it is worth pointing out explicitly how the
-#' accuracy on training set is still greater than on the test set
-#' for the augmented network as well. This is also not a
-#' surprise, given that the augmented images are likely more
-#' similar to their original ones than to the test images.
-
-#' For the baseline network, the accuracy on the test set
-#' plateaus quite quickly (around 90%). For the augmented network
-#' on the other hand, it the accuracy keeps increasing for quite
-#' a while longer.
-
-#' ## References
-#'
-#' [^MNIST1998]: LeCun, Yan, Corinna Cortes, Christopher J.C. Burges. ["The MNIST database of handwritten digits"](http://yann.lecun.com/exdb/mnist/) Website. 1998.
diff --git a/examples/mnist_tensorflow.jl.old b/examples/mnist_tensorflow.jl.old
deleted file mode 100644
index 56f62385..00000000
--- a/examples/mnist_tensorflow.jl.old
+++ /dev/null
@@ -1,463 +0,0 @@
-#' # MNIST: TensorFlow CNN
-info("MNIST: TensorFlow CNN example") #jl-only
-
-#' In this tutorial we will adapt the
-#' [MNIST example](https://github.com/malmaud/TensorFlow.jl/blob/master/examples/mnist_full.jl)
-#' from [TensorFlow.jl](https://github.com/malmaud/TensorFlow.jl)
-#' to utilize a custom augmentation pipeline.
-#' In order to showcase the effect that image augmentation can
-#' have on a neural network's ability to generalize, we will
-#' limit the training set to just the first 500 images (of the
-#' available 60,000!). For more information on the dataset see
-#' [^MNIST1998].
-
-#md #' !!! note
-#md #'
-#md #' This tutorial is also available as a
-#md #' [Juypter](https://jupyter.org/) notebook. You can
-#md #' find a link to the Juypter version of this tutorial
-#md #' in the top right corner of this page.
-
-#' ## Preparing the MNIST dataset
-info("Preparing the MNIST dataset") #jl-only
-
-#' In order to access, prepare, and visualize the MNIST images we
-#' employ the help of three additional Julia packages. In the
-#' interest of time and space we will not go into great detail
-#' about their functionality. Feel free to click on their
-#' respective names to find out more information about the
-#' utility they can provide.
-#'
-#' - [MLDatasets.jl](https://github.com/JuliaML/MLDatasets.jl)
-#' has an MNIST submodule that offers a convenience interface
-#' to read the MNIST database.
-#'
-#' - [Images.jl](https://github.com/JuliaImages/Images.jl) will
-#' provide us with the necessary tools to process and display
-#' the image data in Julia / Juypter.
-#'
-#' - [MLDataUtils.jl](https://github.com/JuliaML/MLDataUtils.jl)
-#' implements a variety of functions to convert and partition
-#' Machine Learning datasets. This will help us prepare the
-#' MNIST data to be used with TensorFlow.
-
-using Images, MLDatasets, MLDataUtils
-srand(42);
-#md nothing # hide
-
-#' As you may have seen previously in the
-#' [elastic distortions tutorial](@ref elastic), the function
-#' `MNIST.traintensor` returns the MNIST training images
-#' corresponding to the given indices as a multi-dimensional
-#' array. These images are stored in the native horizontal-major
-#' memory layout as a single array of `Float64`. All the
-#' individual values are scaled to be between `0.0` and `1.0`.
-#' Also note, how the observations are laid out along the last
-#' array dimension
-
-@show summary(MNIST.traintensor(1:500));
-#md nothing # hide
-
-#' The corresponding label of each image is stored as an integer
-#' value between `0` and `9`. That means that if the label has
-#' the value `3`, then the corresponding image is known to be a
-#' handwritten "3". To show a more concrete example, the
-#' following code reveals that the first training image denotes a
-#' "5" and the second training image a "0" (etc).
-
-@show summary(MNIST.trainlabels(1:500))
-println("First eight labels: ", join(MNIST.trainlabels(1:8),", "))
-
-#' For TensorFlow we will require a slightly different dimension
-#' layout for the images. More specifically, we will move the
-#' observations into the first array dimension. The labels will
-#' be transformed into a one-of-k matrix. For performance reasons,
-#' we will further convert all the numerical values to be of type
-#' `Float32`. We will do all this by creating a little utility
-#' function that we will name `prepare_mnist`.
-
-"""
- prepare_mnist(tensor, labels) -> (X, Y)
-
-Change the dimension layout x1×x2×N of the given array
-`tensor` to N×x1×x2 and store the result in `X`.
-The given vector `labels` is transformed into a 10×N
-one-hot matrix `Y`. Both, `X` and `Y`, will have the
-element type `Float32`.
-"""
-function prepare_mnist(tensor, labels)
- features = convert(Array{Float32}, permutedims(tensor, (3,1,2)))
- targets = convertlabel(LabelEnc.OneOfK{Float32}, labels, 0:9, ObsDim.First())
- features, targets
-end
-#md nothing # hide
-
-#' With `prepare_mnist` defined, we can now use it in conjunction
-#' with the functions in the `MLDatasets.MNIST` sub-module to load
-#' and prepare our training set. Recall that for this tutorial only
-#' use the first 500 images of the training set will be used.
-
-train_x, train_y = prepare_mnist(MNIST.traintensor(1:500), MNIST.trainlabels(1:500))
-@show summary(train_x) summary(train_y);
-[MNIST.convert2image(train_x[i,:,:]) for i in 1:8]
-#md tmp = hcat(ans...) # hide
-#md save("mnist_tf_train.png",repeat(tmp, inner=(4,4))) # hide
-#md nothing # hide
-
-#md #' 
-
-#' Similarly, we use `MNIST.testtensor` and `MNIST.testlabels`
-#' to load the full MNIST test set. We will utilize that data to
-#' measure how well the network is able to generalize with and
-#' without augmentation.
-
-test_x, test_y = prepare_mnist(MNIST.testtensor(), MNIST.testlabels())
-@show summary(test_x) summary(test_y);
-[MNIST.convert2image(test_x[i,:,:]) for i in 1:8]
-#md tmp = hcat(ans...) # hide
-#md save("mnist_tf_test.png",repeat(tmp, inner=(4,4))) # hide
-#md nothing # hide
-
-#md #' 
-
-#' ## Defining the Network
-info("Defining the Network") #jl-only
-
-#' With the dataset prepared, we can now instantiate our neural
-#' network. To keep things simple, we will use the same
-#' convolutional network as defined in the
-#' [MNIST example](https://github.com/malmaud/TensorFlow.jl/blob/master/examples/mnist_full.jl)
-#' of Julia's TensorFlow package.
-
-using TensorFlow, Distributions
-session = Session(Graph());
-#md nothing # hide
-
-#+
-
-function weight_variable(shape...)
- initial = map(Float32, rand(Normal(0, .001), shape...))
- return Variable(initial)
-end
-
-function bias_variable(shape...)
- initial = fill(Float32(.1), shape...)
- return Variable(initial)
-end
-
-function conv2d(x, W)
- nn.conv2d(x, W, [1, 1, 1, 1], "SAME")
-end
-
-function max_pool_2x2(x)
- nn.max_pool(x, [1, 2, 2, 1], [1, 2, 2, 1], "SAME")
-end
-#md nothing # hide
-
-#+
-
-@tf begin
- x = placeholder(Float32)
- y = placeholder(Float32)
-
- W_conv1 = weight_variable(5, 5, 1, 32)
- b_conv1 = bias_variable(32)
-
- x_image = reshape(x, [-1, 28, 28, 1])
-
- h_conv1 = nn.relu(conv2d(x_image, W_conv1) + b_conv1)
- h_pool1 = max_pool_2x2(h_conv1)
-
- W_conv2 = weight_variable(5, 5, 32, 64)
- b_conv2 = bias_variable(64)
-
- h_conv2 = nn.relu(conv2d(h_pool1, W_conv2) + b_conv2)
- h_pool2 = max_pool_2x2(h_conv2)
-
- W_fc1 = weight_variable(7*7*64, 1024)
- b_fc1 = bias_variable(1024)
-
- h_pool2_flat = reshape(h_pool2, [-1, 7*7*64])
- h_fc1 = nn.relu(h_pool2_flat * W_fc1 + b_fc1)
-
- keep_prob = placeholder(Float32)
- h_fc1_drop = nn.dropout(h_fc1, keep_prob)
-
- W_fc2 = weight_variable(1024, 10)
- b_fc2 = bias_variable(10)
-
- y_conv = nn.softmax(h_fc1_drop * W_fc2 + b_fc2)
-
- global cross_entropy = reduce_mean(-reduce_sum(y.*log(y_conv+1e-8), axis=[2]))
- global optimizer = train.minimize(train.AdamOptimizer(1e-4), cross_entropy)
-
- correct_prediction = broadcast(==, indmax(y_conv, 2), indmax(y, 2))
- global accuracy = reduce_mean(cast(correct_prediction, Float32))
-end
-#md nothing # hide
-
-#' ## Training without Augmentation
-info("Training baseline network without augmentation") #jl-only
-
-#' In order to get an intuition for how useful augmentation can
-#' be, we need a sensible baseline to compare to. To that end, we
-#' will first train the network we just defined using only the
-#' (unaltered) 500 training examples.
-
-#' The package
-#' [ValueHistories.jl](https://github.com/JuliaML/ValueHistories.jl)
-#' will help us record the accuracy during the training process.
-#' We will use those logs later to visualize the differences
-#' between having augmentation or no augmentation.
-
-using ValueHistories
-using ProgressMeter #jl-only
-
-#' To keep things simple, we will not overly optimize our
-#' training function. Thus, we will be content with using a
-#' closure. Because both, the baseline and the augmented version,
-#' will share this "inefficiency", we should still get a decent
-#' enough picture of their performance differences.
-
-function train_baseline(; epochs=500, batchsize=100, reset=true)
- reset && run(session, global_variables_initializer())
- log = MVHistory()
- p = Progress(epochs, desc="Baseline: ") #jl-only
- for epoch in 1:epochs
- for (batch_x, batch_y) in eachbatch(shuffleobs((train_x, train_y), obsdim=1), size=batchsize, obsdim=1)
- run(session, optimizer, Dict(x=>batch_x, y=>batch_y, keep_prob=>0.5))
- end
-
- next!(p) #jl-only
- if (epoch % 50) == 0
- train = run(session, accuracy, Dict(x=>train_x, y=>train_y, keep_prob=>1.0))
- test = run(session, accuracy, Dict(x=>test_x, y=>test_y, keep_prob=>1.0))
- @trace log epoch train test
- msg = "epoch " * lpad(epoch,4) * ": train accuracy " * rpad(round(train,3),5,"0") * ", test accuracy " * rpad(round(test,3),5,"0")
- cancel(p, msg, :blue) #jl-only
-#md println(msg)
-#jp println(msg)
- end
- end
- finish!(p) #jl-only
- log
-end
-#md nothing # hide
-
-#' Aside from the accuracy, we will also keep an eye on the
-#' training time. In particular we would like to see if and how
-#' the addition of augmentation causes our training time to
-#' increase.
-
-train_baseline(epochs=1) # warm-up
-baseline_log = @time train_baseline(epochs=1000);
-#md nothing # hide
-
-#' As we can see, the accuracy on the training set is around a
-#' 100%, while the accuracy on the test set peaks around 85%. For
-#' a mere 500 training examples, this isn't actually that bad of
-#' a result.
-
-#' ## Integrating Augmentor
-info("Training network with augmentation") #jl-only
-
-#' Now that we have a network architecture with a baseline to
-#' compare to, let us finally see what it takes to add Augmentor
-#' to our experiment. First, we need to include the package to
-#' our experiment.
-
-using Augmentor
-
-#' The next step, and maybe the most human-hour consuming part of
-#' adding image augmentation to a prediction problem, is to
-#' design and select a sensible augmentation pipeline. Take a
-#' look at the [elastic distortions tutorial](@ref elastic) for
-#' an example of how to do just that.
-
-#' For this example, we already choose a quite complicated but
-#' promising augmentation pipeline for you. This pipeline was
-#' designed to yield a large variation of effects as well as to
-#' showcase how even deep pipelines are quite efficient in terms
-#' of performance.
-
-pl = PermuteDims(2,1) |>
- ShearX(-5:5) * ShearY(-5:5) |>
- Rotate(-15:15) |>
- CropSize(28,28) |>
- Zoom(0.9:0.1:1.2) |>
- CacheImage() |>
- ElasticDistortion(10) |>
- PermuteDims(2,1)
-println(pl) #jl-only
-
-#' Most of the used operations are quite self explanatory, but
-#' there are some details about this pipeline worth pointing out
-#' explicitly.
-
-#' 1. We use the operation [`PermuteDims`](@ref) to convert the
-#' horizontal-major MNIST image to a julia-native
-#' vertical-major image. The vertical-major image is then
-#' processed and converted back to a horizontal-major array.
-#' We mainly do this here to showcase the option, but it is
-#' also to keep consistent with how the data is usually used
-#' in the literature. Alternatively, one could just work with
-#' the MNIST data in a vertical-major format all the way
-#' through without any issue.
-
-#' 2. As counter-intuitive as it sounds, the operation
-#' [`CacheImage`](@ref) right before
-#' [`ElasticDistortion`](@ref) is actually used to improve
-#' performance. If we were to omit it, then the whole pipeline
-#' would be applied in one single pass. In this case, applying
-#' distortions on top of affine transformations lazily is in
-#' fact less efficient than using a temporary variable.
-
-#' With the pipeline now defined, let us quickly peek at what
-#' kind of effects we can achieve with it. In particular, lets
-#' apply the pipeline multiple times to the first training image
-#' and look at what kind of results it produces.
-
-[MNIST.convert2image(augment(train_x[1,:,:], pl)) for i in 1:8, j in 1:2]
-#md tmp = vcat(hcat(ans[:,1]...), hcat(ans[:,2]...)) # hide
-#md save("mnist_tf_aug.png",repeat(tmp, inner=(4,4))) # hide
-#md nothing # hide
-
-#md #' 
-
-#' As we can see, we can achieve a wide range of effects, from
-#' more subtle to more pronounced. The important part is that all
-#' examples are still clearly representative of the true label.
-
-#' Next, we have to adapt the function `train_baseline` to make
-#' use of our augmentation pipeline. To integrate Augmentor
-#' efficiently, there are three necessary changes we have to
-#' make.
-
-#' 1. Preallocate a buffer with the same size and element type
-#' that each batch has.
-#'
-#' ```
-#' augmented_x = zeros(Float32, batchsize, 28, 28)
-#' ```
-
-#' 2. Add a call to [`augmentbatch!`](@ref) in the inner loop of
-#' the batch iterator using our pipeline and buffer.
-#'
-#' ```
-#' augmentbatch!(augmented_x, batch_x, pl, ObsDim.First())
-#' ```
-
-#' 3. Replace `x=>batch_x` with `x=>augmented_x` in the call to
-#' TensorFlow's `run(session, ...)`.
-
-#' Applying these changes to our `train_baseline` function
-#' will give us something similar to the following function.
-#' Note how all the other parts of the function remain exactly
-#' the same as before.
-
-function train_augmented(; epochs=500, batchsize=100, reset=true)
- reset && run(session, global_variables_initializer())
- log = MVHistory()
- p = Progress(epochs, desc="Augmented: ") #jl-only
- augm_x = zeros(Float32, batchsize, size(train_x,2), size(train_x,3))
- for epoch in 1:epochs
- for (batch_x, batch_y) in eachbatch(shuffleobs((train_x, train_y), obsdim=1), size=batchsize, obsdim=1)
- augmentbatch!(CPUThreads(), augm_x, batch_x, pl, ObsDim.First())
- run(session, optimizer, Dict(x=>augm_x, y=>batch_y, keep_prob=>0.5))
- end
-
- next!(p) #jl-only
- if (epoch % 50) == 0
- train = run(session, accuracy, Dict(x=>train_x, y=>train_y, keep_prob=>1.0))
- test = run(session, accuracy, Dict(x=>test_x, y=>test_y, keep_prob=>1.0))
- @trace log epoch train test
- msg = "epoch " * lpad(epoch,4) * ": train accuracy " * rpad(round(train,3),5,"0") * ", test accuracy " * rpad(round(test,3),5,"0")
- cancel(p, msg, :blue) #jl-only
-#md println(msg)
-#jp println(msg)
- end
- end
- finish!(p) #jl-only
- log
-end
-#md nothing # hide
-
-#' You may have noticed in the code above that we also pass a
-#' `CPUThreads()` as the first argument to [`augmentbatch!`](@ref).
-#' This instructs Augmentor to process the images of the batch in
-#' parallel using multi-threading. For this to work properly you
-#' will need to set the environment variable `JULIA_NUM_THREADS`
-#' to the number of threads you wish to use. You can check how
-#' many threads are used with the function `Threads.nthreads()`
-
-@show Threads.nthreads();
-#md nothing # hide
-
-#' Now that all pieces are in place, let us train our network
-#' once more. We will use the same parameters except that now
-#' instead of the original training images we will be using
-#' randomly augmented images. This will cause every epoch to be
-#' different.
-
-train_augmented(epochs=1) # warm-up
-augmented_log = @time train_augmented(epochs=1000);
-#md nothing # hide
-
-#' As we can see, our network reaches far better results on our
-#' testset than our baseline network did. However, we can also
-#' see that the training took quite a bit longer than before.
-#' This difference generally decreases as the complexity of the
-#' utilized neural network increases. Yet another way to improve
-#' performance (aside from simplifying the augmentation pipeline)
-#' would be to increase the number of available threads.
-
-#' ## Visualizing the Results
-info("Visualizing the Results") #jl-only
-
-#' Before we end this tutorial, let us make use the
-#' [Plots.jl](https://github.com/JuliaPlots/Plots.jl) package to
-#' visualize and discuss the recorded training curves.
-#' We will plot the accuracy curves of both networks side by side
-#' in order to get a good feeling about their differences.
-
-using Plots
-#jp pyplot()
-#md pyplot()
-#md nothing # hide
-unicodeplots() #jl-only
-
-#+
-
-#md default(bg_outside=colorant"#FFFFFF") # hide
-plt = plot(
- plot(baseline_log, title="Accuracy (baseline)", ylim=(.5,1)),
- plot(augmented_log, title="Accuracy (augmented)", ylim=(.5,1)),
- size = (900, 400),
- markersize = 1
-)
-#jp plt
-#md png(plt, "mnist_tf_curves.png") # hide
-#md nothing # hide
-
-#md #' 
-
-#' Note how the accuracy on the (unaltered) training set
-#' increases much faster for the baseline network than for the
-#' augmented one. This is to be expected, since our augmented
-#' network doesn't actually use the unaltered images for
-#' training, and thus has not actually seen them. Given this
-#' information, it is worth pointing out explicitly how the
-#' accuracy on training set is still greater than on the test set
-#' for the augmented network as well. This is also not a
-#' surprise, given that the augmented images are likely more
-#' similar to their original ones than to the test images.
-
-#' For the baseline network, the accuracy on the test set
-#' plateaus quite quickly (around 85%). For the augmented network
-#' on the other hand, it the accuracy keeps increasing for quite
-#' a while longer. If you let the network train long enough you
-#' can achieve around 97% even before it stops learning.
-
-#' ## References
-#'
-#' [^MNIST1998]: LeCun, Yan, Corinna Cortes, Christopher J.C. Burges. ["The MNIST database of handwritten digits"](http://yann.lecun.com/exdb/mnist/) Website. 1998.
diff --git a/src/utils.jl b/src/utils.jl
index 0ae90930..03ec4833 100644
--- a/src/utils.jl
+++ b/src/utils.jl
@@ -1,14 +1,26 @@
"""
- testpattern() -> Matrix{RGBA{N0f8}}
+ testpattern([T=RGBA{N0f8}]; ratio=1.0) -> Matrix{RGBA{N0f8}}
-Load and return the provided 300x400 test image.
+Load and return the provided 300x400 test image. Additional args and kwargs
+are passed to `imresize`.
The returned image was specifically designed to be informative
about the effects of the applied augmentation operations. It is
thus well suited to prototype an augmentation pipeline, because it
makes it easy to see what kind of effects one can achieve with it.
"""
-testpattern() = load(joinpath(@__DIR__, "..", "resources", "testpattern.png"))
+function testpattern(args...; ratio=1.0)
+ imresize(load(joinpath(@__DIR__, "..", "resources", "testpattern.png")), ratio=ratio)
+end
+function testpattern(T::Type{<:Colorant}; ratio=1.0)
+ # Directly call T.(testpattern) returns a testpattern with border filled with black pixels
+ # This patch fills border with white pixels so as to be consistent with ARGB(0, 0, 0, 0).
+ npad = 20
+ temp = testpattern()
+ out = fill(oneunit(T), size(temp))
+ out[npad:end-npad, npad:end-npad] .= temp[npad:end-npad, npad:end-npad]
+ return imresize(out, ratio=ratio)
+end
function use_testpattern()
@info("No custom image specifed. Using \"testpattern()\" for demonstration.")